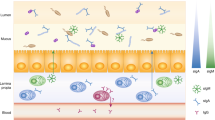
Abstract
Disruptions to the microbiota can have pathological consequences, which highlights the need to understand the factors that contribute to its stability. Although decades of research have focused on the importance of IgA during pathogenic infection, much of the IgA that is generated in the gut targets the resident commensal microorganisms. Despite this observation, the role of antibodies in regulating microbiota composition remains controversial and poorly understood. Here we propose that antibodies generated in response to microbial colonization of the gut shape the composition of the microbiota to benefit the health of the host through a process that we term antibody-mediated immunoselection (AMIS). Given the exquisite specificity of antibodies and an emerging interest in the use of immunotherapies, we suggest that understanding AMIS of the microbiota will highlight novel uses of antibodies to manipulate microbial communities for therapeutic benefit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lindner, C. et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J. Exp. Med. 209, 365–377 (2012).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Yoshida, M. et al. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity 20, 769–783 (2004).
Smith, K., McCoy, K. D. & Macpherson, A. J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69 (2007).
Macpherson, A. J. et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Benveniste, J., Lespinats, G., Adam, C. & Salomon, J. C. Immunoglobulins in intact, immunized, and contaminated axenic mice: study of serum IgA. J. Immunol. 107, 1647–1655 (1971).
Shroff, K. E., Meslin, K. & Cebra, J. J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 (1995).
van der Waaij, L. A., Mesander, G., Limburg, P. C. & van der Waaij, D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry 16, 270–279 (1994).
Kawamoto, S. et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336, 485–489 (2012).
Kubinak, J. L. et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163 (2015).
Kubinak, J. L. et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat. Commun. 6, 8642 (2015).
Bunker, J. J. et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015).
Planer, J. D. et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Kau, A. L. et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl Med. 7, 276ra24 (2015).
Cullender, T. C. et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581 (2013).
Rogier, E. W., Frantz, A. L., Bruno, M. E. & Kaetzel, C. S. Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens 3, 390–403 (2014).
Ermund, A., Gustafsson, J. K., Hansson, G. C. & Keita, A. V. Mucus properties and goblet cell quantification in mouse, rat and human ileal Peyer's patches. PLoS ONE 8, e83688 (2013).
Phalipon, A. et al. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity 17, 107–115 (2002).
Finlay, B. B. & McFadden, G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006).
Dimitriu, P. A. et al. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ. Microbiol. Rep. 5, 200–210 (2013).
Zhang, H., Sparks, J. B., Karyala, S. V., Settlage, R. & Luo, X. M. Host adaptive immunity alters gut microbiota. ISME J. 9, 770–781 (2015).
Kawamoto, S. et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014).
Fagarasan, S., Kinoshita, K., Muramatsu, M., Ikuta, K. & Honjo, T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature 413, 639–643 (2001).
Fagarasan, S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004).
Wei, M. et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 12, 264–270 (2011).
Mirpuri, J. et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5, 28–39 (2014).
Reikvam, D. H. et al. Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. Eur. J. Immunol. 42, 2959–2970 (2012).
Kroese, F. G. et al. Many of the IgA producing plasma cells in murine gut are derived from self-replenishing precursors in the peritoneal cavity. Int. Immunol. 1, 75–84 (1989).
Suzuki, K., Ha, S. A., Tsuji, M. & Fagarasan, S. Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Semin. Immunol. 19, 127–135 (2007).
Fagarasan, S., Kawamoto, S., Kanagawa, O. & Suzuki, K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 (2010).
Fagarasan, S. & Honjo, T. Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol. 3, 63–72 (2003).
Suzuki, K., Kawamoto, S., Maruya, M. & Fagarasan, S. GALT: organization and dynamics leading to IgA synthesis. Adv. Immunol. 107, 153–185 (2010).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Shulzhenko, N. et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 17, 1585–1593 (2011).
Morteau, O. et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J. Immunol. 181, 6309–6315 (2008).
Lindner, C. et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 16, 880–888 (2015).
Rogier, E. W. et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl Acad. Sci. USA 111, 3074–3079 (2014).
Koch, M. A. et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016).
Mikhailov, T. A. & Furner, S. E. Breastfeeding and genetic factors in the etiology of inflammatory bowel disease in children. World J. Gastroenterol. 15, 270–279 (2009).
Hanson, L. A. & Winberg, J. Breast milk and defence against infection in the newborn. Arch. Dis. Child. 47, 845–848 (1972).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Ha, S. A. et al. Regulation of B1 cell migration by signals through Toll-like receptors. J. Exp. Med. 203, 2541–2550 (2006).
Kirkland, D. et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity 36, 228–238 (2012).
He, B. et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol. 11, 836–845 (2010).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Johansen, F. E. et al. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 190, 915–922 (1999).
Zeng, M. Y. et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016).
Lycke, N., Erlandsson, L., Ekman, L., Schon, K. & Leanderson, T. Lack of J chain inhibits the transport of gut IgA and abrogates the development of intestinal antitoxic protection. J. Immunol. 163, 913–919 (1999).
Forbes, S. J., Eschmann, M. & Mantis, N. J. Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun. 76, 4137–4144 (2008).
Mantis, N. J. & Forbes, S. J. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Invest. 39, 383–406 (2010).
Kamada, N. et al. Humoral immunity in the gut selectively targets phenotypically virulent attaching-and-effacing bacteria for intraluminal elimination. Cell Host Microbe 17, 617–627 (2015).
Ruemmele, F. M. et al. Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology 115, 822–829 (1998).
Acknowledgements
The authors would like to thank members of the Round laboratory for their critical review of the manuscript. Support for the laboratory comes from University of Utah's seed grant program, NIH innovator award DP2AT008746-01, Edward Mallinckrodt Jr. Foundation, Pew Scholars Program, NSF CAREER award (IOS-1253278), Packard Fellowship in Science and Engineering and NIAID K22 (AI95375) and NIAID R21 AI109122 to J.L.R.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kubinak, J., Round, J. Do antibodies select a healthy microbiota?. Nat Rev Immunol 16, 767–774 (2016). https://doi.org/10.1038/nri.2016.114
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2016.114
This article is cited by
-
Fecal Immunoglobulin Levels as a Modifier of the Gut Microbiome in Patients with Common Variable Immunodeficiency
Journal of Clinical Immunology (2023)
-
Microbial symphony orchestrated by mucosal IgA
Cellular & Molecular Immunology (2022)
-
Defective humoral immunity disrupts bile acid homeostasis which promotes inflammatory disease of the small bowel
Nature Communications (2022)
-
Diversity and dynamism of IgA−microbiota interactions
Nature Reviews Immunology (2021)
-
NOD2 deficiency increases retrograde transport of secretory IgA complexes in Crohn’s disease
Nature Communications (2021)