Key Points
-
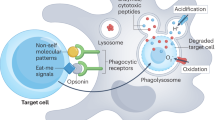
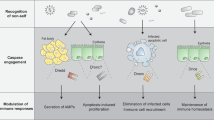
Phagocytosis, the internalization of particles by cells, is involved in development, tissue homeostasis and host defence. Aspects of this process are highly conserved between species and hence studies in model organisms, such as Drosophila melanogaster, have been informative.
-
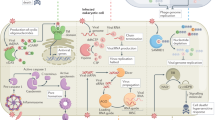
Particle internalization is not simply a mechanism of waste disposal but rather essential for the formation of the phagosome, the organelle that is generated around the internalized particle. As in mammals, D. melanogaster phagosomes are highly complex compartments and are central to many of the effector functions of phagocytes.
-
Phagocytosis is triggered after ligation of cell-surface receptors. Similar to mammals, D. melanogaster uses multiple mechanisms of bacterial recognition, such as complement-like opsonization and scavenger receptors. In addition, studies in D. melanogaster have highlighted a new family of phagocytic receptors that use epidermal growth factor (EGF)-like repeats to bind ligands.
-
Fruit flies have an alternative to the highly variable antibodies found in mammals and instead use DSCAM (Down syndrome cell-adhesion molecule), an immunoglobulin-superfamily member that shows remarkable variability, that may act as both an opsonin and a cell-surface receptor.
-
RNA-interference-based screens have shown that much of the machinery downstream of the cell-surface receptors are common between flies and humans. Studies in D. melanogaster have also suggested roles for the coatamer protein complex and the exocyst complex in the process of phagocytosis.
-
In addition, studies in D. melanogaster have added to our understanding of the role of phagocytosis in pathogen sensing. Furthermore, D. melanogaster phagocytes may potentially be involved in immune adaptation and the processing of internalized antigens.
Abstract
Phagocytosis, the engulfment of material by cells, is a highly conserved process that arose before the development of multicellularity. Phagocytes have a key role in embryogenesis and also guard the portals of potential pathogen entry. They discriminate between diverse particles through the array of receptors expressed on their surface. In higher species, arguably the most sophisticated function of phagocytes is the processing and presentation of antigens derived from internalized material to stimulate lymphocytes and long-lived specific immunity. Central to these processes is the generation of a phagosome, the organelle that forms around internalized material. As we discuss in this Review, over the past two decades important insights into phagocytosis have been gleaned from studies in the model organism Drosophila melanogaster.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chen, G., Zhuchenko, O. & Kuspa, A. Immune-like phagocyte activity in the social amoeba. Science 317, 678–681 (2007).
Tepass, U., Fessler, L. I., Aziz, A. & Hartenstein, V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120, 1829–1837 (1994).
Zhou, L., Hashimi, H., Schwartz, L. M. & Nambu, J. R. Programmed cell death in the Drosophila central nervous system midline. Curr. Biol. 5, 784–790 (1995).
Gordon, S. et al. Localization and function of tissue macrophages. Ciba Found. Symp. 118, 54–67 (1986).
Hopkinson-Woolley, J., Hughes, D., Gordon, S. & Martin, P. Macrophage recruitment during limb development and wound healing in the embryonic and foetal mouse. J. Cell Sci. 107 (Pt 5), 1159–1167 (1994).
Hoffmann, J. A., Kafatos, F. C., Janeway, C. A. & Ezekowitz, R. A. Phylogenetic perspectives in innate immunity. Science 284, 1313–1318 (1999).
Vogel, G., Thilo, L., Schwarz, H. & Steinhart, R. Mechanism of phagocytosis in Dictyostelium discoideum: phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J. Cell Biol. 86, 456–465 (1980).
Ezekowitz, R. A. et al. Uptake of Pneumocystis carinii mediated by the macrophage mannose receptor. Nature 351, 155–158 (1991).
Kruskal, B. A., Sastry, K., Warner, A. B., Mathieu, C. E. & Ezekowitz, R. A. Phagocytic chimeric receptors require both transmembrane and cytoplasmic domains from the mannose receptor. J. Exp. Med. 176, 1673–1680 (1992).
Aderem, A. & Underhill, D. M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17, 593–623 (1999).
Greenberg, S. & Grinstein, S. Phagocytosis and innate immunity. Curr. Opin. Immunol. 14, 136–145 (2002).
Cardelli, J. Phagocytosis and macropinocytosis in Dictyostelium: phosphoinositide-based processes, biochemically distinct. Traffic 2, 311–320 (2001).
Gumienny, T. L. & Hengartner, M. O. How the worm removes corpses: the nematode C. elegans as a model system to study engulfment. Cell Death Differ. 8, 564–568 (2001).
Franc, N. C., Heitzler, P., Ezekowitz, R. A. & White, K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284, 1991–1994 (1999). This study shows that Croquemort is required for efficient phagocytosis of apopototic cell corpses in the D. melanogaster embryo in vivo and that its expression level is regulated by the amount of apoptosis in the embryo.
Pearson, A. M. et al. Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect. 5, 815–824 (2003). This study describes the use of a genetic-deficiency screen in primary larval haemocytes ex vivo to identify several cytoskeletal proteins that are involved in phagocytosis. It demonstrates the use of D. melanogaster to address molecular mechanisms underlying bacterial phagocytosis and also establishes the validity of using S2 cells for this purpose.
Silva, E., Au-Yeung, H. W., Van Goethem, E., Burden, J. & Franc, N. C. Requirement for a Drosophila E3-ubiquitin ligase in phagocytosis of apoptotic cells. Immunity 27, 585–596 (2007).
Ramet, M. et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity 15, 1027–1038 (2001). In this study, compounds related to scavenger-receptor ligands were used to inhibit phagocytosis of bacteria by S2 cells and led to the identification of a D. melanogaster scavenger receptor that is involved in this process.
Rabinowitz, S., Horstmann, H., Gordon, S. & Griffiths, G. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J. Cell Biol. 116, 95–112 (1992).
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002). This reference describes the first high-throughput RNAi screen in S2 cells, identifying roles for the GATA transcription factor Serpent, COPI and COPII proteins and a member of the PGRP family (PGRP-LC) in bacterial phagocytosis in D. melanogaster.
Stuart, L. M. et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 170, 477–485 (2005). This study describes the use of a high-throughput RNAi screen in S2 cells that led to the investigation of the role of mammalian CD36 in the phagocytosis and response to S. aureus infection.
Stuart, L. M. et al. A systems biology analysis of the Drosophila phagosome. Nature 445, 95–101 (2007). This paper describes a systems-biology approach combining proteomics, bioinformatics and high-throughput RNAi that provided the first comprehensive analysis of phagosomes. It highlights novel and unexpected roles in phagocytosis for COPI and COPII and exocyst complexes.
Philips, J. A., Rubin, E. J. & Perrimon, N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309, 1251–1253 (2005). A genome-wide RNAi screen in S2 cells identified the D. melanogaster gene peste , a CD36 family member, and paved the way for the finding that CD36 family members serve as receptors for mycobacterial entry into human cells.
Stroschein-Stevenson, S. L., Foley, E., O' Farrell P, H. & Johnson, A. D. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4, e4 (2005). This study describes a genome-wide RNAi screen for evolutionarily conserved D. melanogaster genes involved in the recognition and phagocytosis of C. albicans that establishes various TEPs as microbial opsonins that can distinguish C. albicans (MCR), E. coli (TEPII) and S. aureus (TEPIII).
Agaisse, H. et al. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309, 1248–1251 (2005). This study describes the use of a genome-wide RNAi screen in S2 cells to address commonalities and differences in host cell invasion and intracellular growth by bacteria with different cellular replication sites.
Cheng, L. W. et al. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc. Natl Acad. Sci. USA 102, 13646–13651 (2005). In this study three genome-wide RNAi screens in S2 cells were used to elucidate host molecules involved in different facets of cell parasitism (entry, vacuolar escape and intracellular growth) by the bacterial pathogen L. monocytogenes.
Ayres, J. S. & Schneider, D. S. Genomic dissection of microbial pathogenesis in cultured Drosophila cells. Trends Microbiol. 14, 101–104 (2006).
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007).
Brennan, C. A. & Anderson, K. V. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22, 457–483 (2004).
Elrod-Erickson, M., Mishra, S. & Schneider, D. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 10, 781–784 (2000). In this study functional ablation of phagocytes by polystyrene beads was used to demonstrate that humoral defence mechanisms act together with phagocytosis to generate effective immune responses in D. melanogaster.
Kocks, C. et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123, 335–346 (2005). This study establishes that phagocytosis is a cellular host defence mechanism in D. melanogaster . Transcriptional profiling and high-throughput RNAi in S2 cells was used to identify a new type of EGF-like-repeat-containing receptor (Eater) that has a crucial role in the host defence against bacterial infections in vivo.
Williams, M. J., Wiklund, M. L., Wikman, S. & Hultmark, D. Rac1 signalling in the Drosophila larval cellular immune response. J. Cell Sci. 119, 2015–2024 (2006).
Williams, M. J., Ando, I. & Hultmark, D. Drosophila melanogaster Rac2 is necessary for a proper cellular immune response. Genes Cells 10, 813–823 (2005).
Avet-Rochex, A., Perrin, J., Bergeret, E. & Fauvarque, M. O. Rac2 is a major actor of Drosophila resistance to Pseudomonas aeruginosa acting in phagocytic cells. Genes Cells 12, 1193–1204 (2007).
Nehme, N. T. et al. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3, e173 (2007).
Garin, J. et al. The phagosome proteome: insight into phagosome functions. J. Cell Biol. 152, 165–180 (2001).
Desjardins, M. ER-mediated phagocytosis: a new membrane for new functions. Nature Rev. Immunol. 3, 280–291 (2003).
Desjardins, M., Huber, L. A., Parton, R. G. & Griffiths, G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124, 677–688 (1994).
Meresse, S. et al. Controlling the maturation of pathogen-containing vacuoles: a matter of life and death. Nature Cell Biol. 1, E183–188 (1999).
Lagueux, M., Perrodou, E., Levashina, E. A., Capovilla, M. & Hoffmann, J. A. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc. Natl Acad. Sci. USA 97, 11427–11432 (2000).
Moita, L. F. et al. In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity 23, 65–73 (2005).
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Gordon, S. Pattern recognition receptors: doubling up for the innate immune response. Cell 111, 927–930 (2002).
Wright, S. D., Ramos, R. A., Tobias, P. S., Ulevitch, R. J. & Mathison, J. C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 (1990).
Hoebe, K. et al. CD36 is a sensor of diacylglycerides. Nature 433, 523–527 (2005).
Savill, J., Hogg, N., Ren, Y. & Haslett, C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Invest. 90, 1513–1522 (1992).
Franc, N. C., Dimarcq, J. L., Lagueux, M., Hoffmann, J. & Ezekowitz, R. A. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4, 431–443 (1996). This paper describes the D. melanogaster receptor Croquemort, a scavenger receptor that is related to mammalian CD36, and shows that it binds to and mediates the uptake of apoptotic cells.
Pearson, A., Lux, A. & Krieger, M. Expression cloning of dSR-CI, a class C macrophage-specific scavenger receptor from Drosophila melanogaster. Proc. Natl Acad. Sci. USA 92, 4056–4060 (1995).
Irving, P., Troxler, L. & Hetru, C. Is innate enough? The innate immune response in Drosophila. C. R. Biol. 327, 557–570 (2004).
Lazzaro, B. P. Elevated polymorphism and divergence in the class C scavenger receptors of Drosophila melanogaster and D. simulans. Genetics 169, 2023–2034 (2005).
Kurucz, E. et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17, 649–654 (2007).
Ju, J. S. et al. A novel 40-kDa protein containing six repeats of an epidermal growth factor-like domain functions as a pattern recognition protein for lipopolysaccharide. J. Immunol. 177, 1838–1845 (2006).
Freeman, M. R., Delrow, J., Kim, J., Johnson, E. & Doe, C. Q. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38, 567–580 (2003).
Manaka, J. et al. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J. Biol. Chem. 279, 48466–48476 (2004).
Awasaki, T. et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 (2006).
MacDonald, J. M. et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50, 869–881 (2006).
Su, H. P. et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J. Biol. Chem. 277, 11772–11779 (2002).
Hamon, Y. et al. Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS ONE 1, e120 (2006).
Schmucker, D. & Flanagan, J. G. Generation of recognition diversity in the nervous system. Neuron 44, 219–222 (2004).
Schmucker, D. et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000).
Watson, F. L. et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309, 1874–1878 (2005). This paper describes the hypervariable, neuronal immunoglobulin-superfamily receptor DSCAM as being abundantly expressed in the immune tissues of D. melanogaster and implicates thousands of alternative splice forms as PRRs and opsonins in the phagocytosis of microorganisms.
Meijers, R. et al. Structural basis of Dscam isoform specificity. Nature 449, 487–491 (2007).
Neves, G., Zucker, J., Daly, M. & Chess, A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 36, 240–246 (2004).
Dong, Y., Taylor, H. E. & Dimopoulos, G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 4, e229 (2006).
Pancer, Z. et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 (2004).
Caron, E. & Hall, A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282, 1717–1721 (1998).
Hall, A. B. et al. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcγR- and complement-mediated phagocytosis. Immunity 24, 305–316 (2006).
Olazabal, I. M. et al. Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcγR, phagocytosis. Curr. Biol. 12, 1413–1418 (2002).
Colucci-Guyon, E. et al. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. 15, 2007–2012 (2005).
Castellano, F., Chavrier, P. & Caron, E. Actin dynamics during phagocytosis. Semin. Immunol. 13, 347–355 (2001).
Etienne-Manneville, S. & Hall, A. Rho GTPases in cell biology. Nature 420, 629–635 (2002).
Swanson, J. A. & Hoppe, A. D. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 76, 1093–1103 (2004).
Antonny, B. Membrane deformation by protein coats. Curr. Opin. Cell Biol 18, 386–394 (2006).
Gurkan, C., Stagg, S. M., Lapointe, P. & Balch, W. E. The COPII cage: unifying principles of vesicle coat assembly. Nature Rev. Mol. Cell Biol. 7, 727–738 (2006).
Lippincott-Schwartz, J. & Liu, W. Insights into COPI coat assembly and function in living cells. Trends Cell Biol. 16, e1–e4 (2006).
Veiga, E. & Cossart, P. The role of clathrin-dependent endocytosis in bacterial internalization. Trends Cell Biol. 16, 499–504 (2006).
Botelho, R. J., Hackam, D. J., Schreiber, A. D. & Grinstein, S. Role of COPI in phagosome maturation. J. Biol. Chem. 275, 15717–15727 (2000).
TerBush, D. R., Maurice, T., Roth, D. & Novick, P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15, 6483–6494 (1996).
Boyd, C., Hughes, T., Pypaert, M. & Novick, P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167, 889–901 (2004).
Clandinin, T. R. Surprising twists to exocyst function. Neuron 46, 164–166 (2005).
Brennan, C. A., Delaney, J. R., Schneider, D. S. & Anderson, K. V. Psidin is required in Drosophila blood cells for both phagocytic degradation and immune activation of the fat body. Curr. Biol. 17, 67–72 (2007).
Herskovits, A. A., Auerbuch, V. & Portnoy, D. A. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 3, e51 (2007).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Ogura, Y. et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276, 4812–4818 (2001).
Inohara, N., Ogura, Y., Chen, F. F., Muto, A. & Nunez, G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J. Biol. Chem. 276, 2551–2554 (2001).
Kaneko, T. et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nature Immunol. 7, 715–723 (2006).
Takehana, A. et al. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23, 4690–4700 (2004).
Hauton, C. & Smith, V. J. Adaptive immunity in invertebrates: a straw house without a mechanistic foundation. Bioessays 29, 1138–1146 (2007).
Boman, H. G., Nilsson, I. & Rasmuson, B. Inducible antibacterial defence system in Drosophila. Nature 237, 232–235 (1972).
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M. & Schneider, D. S. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 3, e26 (2007).
Kovacsovics-Bankowski, M. & Rock, K. L. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science 267, 243–246 (1995).
Kovacsovics-Bankowski, M., Clark, K., Benacerraf, B. & Rock, K. L. Efficient major histocompatibility complex class I presentation of exogenous antigen upon phagocytosis by macrophages. Proc. Natl Acad. Sci. USA 90, 4942–4946 (1993).
Houde, M. et al. Phagosomes are competent organelles for antigen cross-presentation. Nature 425, 402–406 (2003).
Ackerman, A. L., Kyritsis, C., Tampe, R. & Cresswell, P. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc. Natl Acad. Sci. USA 100, 12889–12894 (2003).
Guermonprez, P. et al. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 (2003).
Touret, N. et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell 123, 157–170 (2005).
Desjardins, M., Houde, M. & Gagnon, E. Phagocytosis: the convoluted way from nutrition to adaptive immunity. Immunol. Rev. 207, 158–165 (2005).
Metchnikoff, I. I. On the present state of the question of immunity in infectious disease [online] (1908).
Wood, W. et al. Wound healing recapitulates morphogenesis in Drosophila embryos. Nature Cell Biol. 4, 907–912 (2002).
Stramer, B. et al. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168, 567–573 (2005).
Rizki, T. M. & Rizki, R. M. The cellular defense system of Drosophila melanogaster. (eds King, R. C. & Akai, H.) 579–604 (Plenum Publishing Corporation, New York, 1984). An insightful book chapter with stunning electron micrographs describing D. melanogaster blood cells and their possible roles in pathogen defence.
Callebaut, I., Mignotte, V., Souchet, M. & Mornon, J. P. EMI domains are widespread and reveal the probable orthologs of the Caenorhabditis elegans CED-1 protein. Biochem. Biophys. Res. Commun. 300, 619–623 (2003).
Acknowledgements
We thank members of the laboratory of Developmental Immunology, Massachusetts General Hospital, for continuous inspiration. Our particular thanks go to C. Kocks for help with the original figure 3 and for critical reading and helpful input into the manuscript. Finally, we apologize to our colleagues whose work we have been unable to cite or discuss owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Phagocytes
-
Cells such as macrophages and neutrophils that internalize particulate material in an actin-dependent manner. Derived from the greek phagein (to eat) and kytos (cell).
- Apoptotic cell
-
A cell that has died by a genetically regulated programme of cell death. It is characterized by cell shrinkage, chromatin condensation, cell-membrane blebbing and DNA fragmentation. Eventually, the cell breaks up into many membrane-bound apoptotic bodies, which are phagocytosed by neighbouring cells.
- Holometabolous insects
-
Insects, such as Drosophila melanogaster, that have a pupal stage during which they undergo complete reorganization and metamorphosis.
- Haemocytes
-
Cells found within the haemolymph of an insect that are equivalent to the blood cells in vertebrates.
- RNA interference
-
The silencing of gene expression by the introduction of double-stranded RNAs that trigger the specific degradation of a homologous target mRNA and often subsequently decrease production of the encoded protein.
- Opsonin
-
A soluble molecule that, when bound to a particle, enhances uptake of the particle by phagocytosis.
- Pattern-recognition receptors
-
Receptors that recognize molecular patterns often associated with pathogens, such as lipopolysaccharide found in the bacterial cell walls of Gram-negative microorganisms. They also recognize molecular patterns expressed by non-pathogenic bacteria.
- N-ethyl-N-nitrosourea (ENU) mutagenesis
-
An alkylating agent and potent mutagen that when given to mice efficiently induces random mutations.
- Gene rearrangement
-
The ordered rearrangement of variable regions of genes encoding antigen receptors that contributes to increase receptor diversity.
- Somatic hypermutation
-
The process by which antigen-activated B cells in germinal centres mutate their rearranged immunoglobulin genes. The B cells are subsequently selected for those expressing the 'best' mutations on the basis of the ability of the surface immunoglobulin to bind antigen.
- Convergent evolution
-
The development of similar characteristics in organisms that are unrelated as a consequence of how each adapts to a similar evolutionary pressure but which occurs after evolutionary divergence.
- Pseudopod
-
A transient protrusion from the cell during cell movement or to envelop a particle that is to be internalized.
- Exocytosis
-
The release of material contained within vesicles by fusion of the vesicles with the plasma membrane.
Rights and permissions
About this article
Cite this article
Stuart, L., Ezekowitz, R. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol 8, 131–141 (2008). https://doi.org/10.1038/nri2240
Issue Date:
DOI: https://doi.org/10.1038/nri2240
This article is cited by
-
Construction of L-type lectin displaying Saccharomyces cerevisiae for Vibrio parahaemolyticus agglutination
International Microbiology (2023)
-
A near-infrared plasma membrane-specific AIE probe for fluorescence lifetime imaging of phagocytosis
Science China Chemistry (2022)
-
Sensing microbial infections in the Drosophila melanogaster genetic model organism
Immunogenetics (2022)
-
Transcriptomic analysis of Macrobrachium rosenbergii (giant fresh water prawn) post-larvae in response to M. rosenbergii nodavirus (MrNV) infection: de novo assembly and functional annotation
BMC Genomics (2019)
-
Innate immunity in the simplest animals – placozoans
BMC Genomics (2019)