Abstract
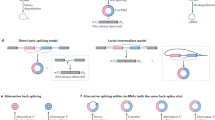
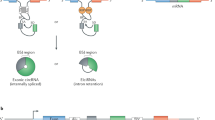
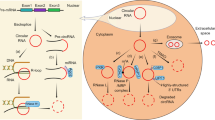
Circular RNAs (circRNAs) are produced from precursor mRNA (pre-mRNA) back-splicing of thousands of genes in eukaryotes. Although circRNAs are generally expressed at low levels, recent findings have shed new light on their cell type-specific and tissue-specific expression and on the regulation of their biogenesis. Furthermore, the data indicate that circRNAs shape gene expression by titrating microRNAs, regulating transcription and interfering with splicing, thus effectively expanding the diversity and complexity of eukaryotic transcriptomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Nigro, J. M. et al. Scrambled exons. Cell 64, 607–613 (1991).
Cocquerelle, C., Daubersies, P., Majerus, M. A., Kerckaert, J. P. & Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11, 1095–1098 (1992).
Capel, B. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030 (1993).
Cocquerelle, C., Mascrez, B., Hetuin, D. & Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160 (1993).
Pasman, Z., Been, M. D. & Garcia-Blanco, M. A. Exon circularization in mammalian nuclear extracts. RNA 2, 603–610 (1996).
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733 (2012).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Guo, J. U., Agarwal, V., Guo, H. & Bartel, D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15, 409 (2014).
Westholm, J. O. et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980 (2014).
Ivanov, A. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 (2015).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Kelly, S., Greenman, C., Cook, P. R. & Papantonis, A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 427, 2414–2417 (2015).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Liang, D. & Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247 (2014).
Starke, S. et al. Exon circularization requires canonical splice signals. Cell Rep. 10, 103–111 (2015).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Kramer, M. C. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29, 2168–2182 (2015).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Zhang, Y. et al. Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806 (2013).
Li, Z. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015).
Lukiw, W. J. Circular RNA (circRNA) in Alzheimer's disease (AD). Front. Genet. 4, 307 (2013).
Burd, C. E. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, e1001233 (2010).
Yang, L., Duff, M. O., Graveley, B. R., Carmichael, G. G. & Chen, L. L. Genome-wide characterization of non-polyadenylated RNAs. Genome Biol. 12, R16 (2011).
Lu, T. et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 21, 2076–2087 (2015).
Wang, P. L. et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9, e90859 (2014).
Barrett, S. P., Wang, P. L. & Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4, e07540 (2015).
Broadbent, K. M. et al. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 16, 454 (2015).
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. & Brown, P. O. Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777 (2013).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015).
You, X. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610 (2015).
Wang, Y. & Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179 (2015).
Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014).
Szabo, L. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16, 126 (2015).
Chen, T. et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 25, 459–476 (2015).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Chen, C. Y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417 (1995).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Denzler, R., Agarwal, V., Stefano, J., Bartel, D. P. & Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 (2014).
Braunschweig, U. et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24, 1774–1786 (2014).
Gardner, E. J., Nizami, Z. F., Talbot, C. C. Jr & Gall, J. G. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 26, 2550–2559 (2012).
Talhouarne, G. J. & Gall, J. G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 20, 1476–1487 (2014).
Armakola, M. et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 44, 1302–1309 (2012).
Bahn, J. H. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61, 221–230 (2015).
Memczak, S., Papavasileiou, P., Peters, O. & Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10, e0141214 (2015).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA 73, 3852–3856 (1976).
Kos, A., Dijkema, R., Arnberg, A. C., van der Meide, P. H. & Schellekens, H. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323, 558–560 (1986).
Flores, R. et al. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 8, 200–206 (2011).
Cote, F. & Perreault, J. P. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 273, 533–543 (1997).
Tang, T. H. et al. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30, 921–930 (2002).
Danan, M., Schwartz, S., Edelheit, S. & Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40, 3131–3142 (2012).
Soma, A. et al. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318, 450–453 (2007).
Grabowski, P. J., Zaug, A. J. & Cech, T. R. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell 23, 467–476 (1981).
Cech, T. R. Self-splicing of group I introns. Annu. Rev. Biochem. 59, 543–568 (1990).
Li-Pook-Than, J. & Bonen, L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 34, 2782–2790 (2006).
Molina-Sanchez, M. D., Martinez-Abarca, F. & Toro, N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 281, 28737–28744 (2006).
Abelson, J., Trotta, C. R. & Li, H. tRNA splicing. J. Biol. Chem. 273, 12685–12688 (1998).
Salgia, S. R., Singh, S. K., Gurha, P. & Gupta, R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 9, 319–330 (2003).
Lu, Z. et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 21, 1554–1565 (2015).
Bachmayr-Heyda, A. et al. Correlation of circular RNA abundance with proliferation—exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 5, 8057 (2015).
Yap, K. L. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 (2010).
Acknowledgements
The author is grateful to Li Yang for discussions. This work was supported by grants 91440202 and 31322018 from the Natural Science Foundation of China (NSFC), and XDA01010206 from the Chinese Academy of Sciences (CAS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Chen, LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17, 205–211 (2016). https://doi.org/10.1038/nrm.2015.32
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2015.32
This article is cited by
-
IGF2BP2-m6A-circMMP9 axis recruits ETS1 to promote TRIM59 transcription in laryngeal squamous cell carcinoma
Scientific Reports (2024)
-
Alzheimer’s Disease-Related Epigenetic Changes: Novel Therapeutic Targets
Molecular Neurobiology (2024)
-
Biological role and regulation of circular RNA as an emerging biomarker and potential therapeutic target for cancer
Molecular Biology Reports (2024)
-
CircSCUBE3 Reduces the Anti-gastric Cancer Activity of Anti-PD-L1
Molecular Biotechnology (2024)
-
Circ_0001944 depletion inhibits glycolysis and esophageal cancer progression by binding to miR-338-5p to reduce PDK1 expression
Journal of Bioenergetics and Biomembranes (2024)