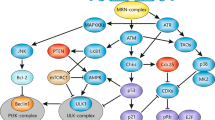
Key Points
-
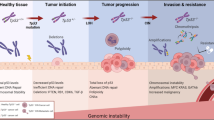
Genomic instability can be linked to disabled apoptosis, and both phenomena can engage in a positive amplification loop.
-
Genomic instability can disable the apoptotic programme — for example, due to the mutation or deletion of pro-apoptotic genes.
-
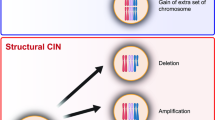
The inhibition of apoptosis can favour chromosomal instability (CIN) at several levels. DNA double-strand breaks result in structural CIN when the default pathway that leads to senescence and apoptosis is blocked. Similarly, telomere dysfunction entails rampant structural CIN only when the p53-dependent senescence or apoptosis pathway is disabled.
-
Inactivation of p53 is also permissive for the survival of polyploid cells. Disabling the DNA-structure checkpoint can favour metaphase-associated cell death. Suppression of this 'mitotic catastrophe' by caspase inhibitors or BCL2 overexpression results in asymmetric cell division and aneuploidy. Furthermore, the spindle-assembly checkpoint is functionally linked to apoptosis regulation by BUBR1 (pro-apoptotic) and survivin (anti-apoptotic) proteins.
-
Oncogenic kinases can simultaneously inhibit DNA repair and apoptosis. Moreover, proteins that are involved in DNA repair can participate in the activation of the DNA-damage-induced apoptotic default pathway or, vice versa, apoptosis-regulatory proteins can affect DNA repair.
-
The intricate interplay between apoptosis control and genomic instability is likely to have an important role in oncogenesis. Proteins that determine this interplay include p53, p21 and CHK2, as well as numerous additional effectors that are involved in the DNA-damage and spindle-assembly checkpoints. Such proteins, as well as their immediate afferent and efferent interactors, might constitute therapeutic targets for cancer prevention and treatment.
Abstract
Genomic instability is intrinsically linked to significant alterations in apoptosis control. Chromosomal and microsatellite instability can cause the inactivation of pro-apoptotic pathways. In addition, the inhibition of apoptosis itself can be permissive for the survival and ongoing division of cells that have failed to repair DNA double-strand breaks, experience telomere dysfunction or are in an abnormal polyploid state. Furthermore, DNA-repair proteins can regulate apoptosis. So, genomic instability and apoptosis are intimately linked phenomena, with important implications for the pathophysiology of cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). Summarizes essential alterations in cell physiology, which dictate malignant transformation.
Pihan, G. & Doxsey, S. J. Mutations and aneuploidy: co-conspirators in cancer? Cancer Cell 4, 89–94 (2003).
Ionov, Y., Yamamoto, H., Krajewski, S., Reed, J. C. & Perucho, M. Mutational inactivation of the proapoptotic gene BAX confers selective advantage during tumor clonal evolution. Proc. Natl Acad. Sci. USA 97, 10872–10877 (2000). First evidence that genomic instability of the microsatellite type can induce mutations in apoptosis-regulatory genes, thereby providing a growth advantage to tumours in vivo . This study is backed up by clinical data that validates the concept that BAX is frequently mutated in colon carcinomas.
Gourdier, I. et al. Drug specific resistance to oxaliplatin is associated with apoptosis defect in a cellular model of colon carcinoma. FEBS Lett. 529, 232–236 (2002).
Reid, B. J. et al. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am. J. Gastroenterol. 96, 2839–2848 (2001).
Sugai, T. et al. Analysis of genetic alterations, classified according to their DNA ploidy pattern, in the progression of colorectal adenomas and early colorectal carcinomas. J. Pathol. 200, 168–176 (2003).
Norbury, C. J. & Zhitovovsky, B. DNA damage-induced apoptosis. Oncogene 23, 2797–2808 (2004).
Shiloh, Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer 3, 155–168 (2003).
Bermudez, V. P. et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA 100, 1633–1638 (2003).
Motoyama, N. & Naka, K. DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev. 14, 11–16 (2004).
Peters, M. et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc. Natl Acad. Sci. USA 99, 11305–11310 (2002).
Takai, H. et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21, 5195–5205 (2002).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002). Pioneering work in which the role for histone H2AX in genomic instability was first proposed.
Taneja, N. et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J. Biol. Chem. 279, 2273–2280 (2004).
Suzuki, K., Yokoyama, S., Waseda, S., Kodama, S. & Watanabe, M. Delayed reactivation of p53 in the progeny of cells surviving ionizing radiation. Cancer Res. 63, 936–941 (2003).
Vousden, K. H. & Lu, X. Live or let die: the cell's response to p53. Nature Rev. Cancer 2, 594–604 (2002).
Konishi, A. et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114, 673–688 (2003).
Chipuk, J. E., Maurer, U., Green, D. R. & Schuler, M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 4, 371–381 (2003). Shows convincing evidence that p53-dependent apoptosis might proceed in a transcriptionally independent manner.
Marchenko, N. D., Zaika, A. & Moll, U. M. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212 (2000).
Mihara, M. et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11, 577–590 (2003). References 19 and 20 show the possible transcription-independent effect of p53 on mitochondria, whereby p53 would interact with BCL-X L.
Chipuk, J. E. et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014 (2004).
Leu, J. I. -J., Dumont, P., Hafey, M., Murphy, M. E. & George, D. L. Mitochondrial p53 activates BAK and leads to disruption of a BAK–MCL1 complex. Nature Cell Biol. 6, 443–450 (2004). References 21 and 22 indicate that the true mitochondrial target of p53 would be BAX (which would be activated in a hit-and-run fashion) or MCL1, respectively.
Melino, G. et al. p73 induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 279, 8076–8083 (2004).
Lin, B. et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 116, 527–540 (2004).
Tinel, A. & Tschopp, J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 (2004). The description of a nuclear caspase-activation complex — the PIDDosome — that contains PIDD (which is also a p53 target gene), the adaptor protein RAIDD/CRAIDD and pro-caspase-2.
Ben-Porath, I. & Weinberg, R. A. When cells get stressed: an integrative view of cellular senescence. J. Clin. Invest. 113, 8–13 (2004).
Artandi, S. E. et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc. Natl Acad. Sci. USA 99, 8191–8196 (2002).
Gisselsson, D. Chromosome instability in cancer: how, when, and why? Adv. Cancer Res. 87, 1–29 (2003).
DePinho, R. A. The age of cancer. Nature 408, 248–254 (2000). Proposes the theory that carcinogenesis is first triggered by a crisis with telomere dysfuction and genomic instability, followed by re-expression of telomerase.
Wong, K. K. et al. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature 421, 643–648 (2003). Reports in vivo evidence that telomere dysfunction, AMT and p53 participate in the regulation of CIN, for example, in the crypts of colon epithelia where cancer stem cells are expected to form.
Chin, L. et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538 (1999).
Zhang, P., Chan, S. L., Fu, W., Mendoza, M. & Mattson, M. P. TERT suppresses apoptotis at a premitochondrial step by a mechanism requiring reverse transcriptase activity and 14-3-3 protein-binding ability. FASEB J. 17, 767–769 (2003).
Kang, H. J. et al. Ectopic expression of the catalytic subunit of telomerase protects against brain injury resulting from ischemia and NMDA-induced neurotoxicity. J. Neurosci. 24, 1280–1287 (2004). References 32 and 33 suggest an apoptosis-inhibitory function of the protein subunit of telomerase that might be dissociated from its effect on telomeres.
Storchova, Z. & Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nature Rev. Mol. Cell Biol. 5, 45–54 (2004).
Raslova, H. et al. Megakaryocyte polyploidization is associated with functional gene amplification. Blood 101, 541–544 (2003).
Guidotti, J. E. et al. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J. Biol. Chem. 278, 19095–19101 (2003).
Mantel, C. et al. p21cip-1/waf-1 deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood 93, 1390–1398 (1999).
Lanni, J. S. & Jacks, T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell Biol. 18, 1055–1064 (1998).
Andreassen, P. R., Lacroix, F. B., Lohez, O. D. & Margolis, R. L. Neither p21WAF1 nor 14-3-3σ prevents G2 progression to mitotic catastrophe in human colon carcinoma cells after DNA damage, but p21WAF1 induces stable G1 arrest in resulting tetraploid cells. Cancer Res. 61, 7660–7668 (2001).
Minn, A. J., Boise, L. H. & Thompson, C. B. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10, 2621–2631 (1996). Established the role of p53 and BCL2-family proteins in the regulation of the cell-cycle checkpoint following the induction of damage to the mitotic spindle.
Chen, J. G., Yang, C. P., Cammer, M. & Horwitz, S. B. Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res. 63, 7891–7899 (2003).
Beltrami, E., Plescia, J., Wilkinson, J. C., Duckett, C. S. & Altieri, D. C. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J. Biol. Chem. 279, 2077–2084 (2004).
Verdoodt, B. et al. Induction of polyploidy and apoptosis after exposure to high concentrations of the spindle poison nocodazole. Mutagenesis 14, 513–520 (1999).
Castedo, M. et al. Human immunodeficiency virus 1 envelope glycoprotein complex-induced apoptosis involves mammalian target of rapamycin/FKBP12-rapamycin-associated protein-mediated p53 phosphorylation. J. Exp. Med. 194, 1097–1110 (2001).
Meraldi, P., Honda, R. & Nigg, E. A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. Embo J. 21, 483–492 (2002). A seminal report that outlines how centrosome amplification could lead to aneuploidy in a p53 -negative context.
Carmena, M. & Earnshaw, W. C. The cellular geography of aurora kinases. Nature Rev. Mol. Cell Biol. 4, 842–854 (2003).
Verschuren, E. W., Klefstrom, J., Evan, G. I. & Jones, N. The oncogenic potential of Kaposi's sarcoma-associated herpesvirus cyclin is exposed by p53 loss in vitro and in vivo. Cancer Cell 2, 229–241 (2002).
Colombel, M. et al. Androgen suppressed apoptosis is modified in p53 deficient mice. Oncogene 10, 1269–1274 (1995).
Ramel, S. et al. Inactivation of p53 and the development of tetraploidy in the elastase-SV40 T antigen transgenic mouse pancreas. Pancreas 11, 213–222 (1995).
Gillet, R. et al. The consequence of p53 overexpression for liver tumor development and the response of transformed murine hepatocytes to genotoxic agents. Oncogene 19, 3498–3507 (2000).
Blount, P. L. et al. 17p allelic losses in diploid cells of patients with Barrett's esophagus who develop aneuploidy. Cancer Res. 54, 2292–2295 (1994).
Castedo, M. et al. Chk2 is a negative regulator of mitotic catastrophe. Oncogene 23, 4353–4361 (2004).
Roninson, I. B., Broude, E. V. & Chang, B. -D. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist. Updat. 4, 303–313 (2001).
Castedo, M., Perfettini, J. -L., Medema, J. P. & Kroemer, G. Cell death by mitotic catastrophe: a molecular definition. Oncogene 23, 2825–2837 (2004).
Castedo, M. et al. Mitotic catastrophe. A special case of apoptosis preventing aneuploidy. Oncogene 23, 4362–4370 (2004). The first systematic investigation of biochemical and morphological events that are related to mitotic catastrophe.
Jallepalli, P. V. & Lengaue, C. Chromosome segregation and cancer: cutting through the mystery. Nature Rev. Cancer 1, 109–117 (2001).
Shin, H. J. et al. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 4, 483–497 (2003).
Lens, S. M. et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22, 2934–2947 (2003). Outlines the role of survivin in the tension arm of the spindle checkpoint.
Liu, T., Brouha, B. & Grossman, D. Rapid induction of mitochondrial events and caspase-independent apoptosis in Survivin-targeted melanoma cells. Oncogene 23, 39–48 (2004).
Song, Z., Yao, X. & Wu, M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 278, 23130–23140 (2003).
Marusawa, H. et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22, 2729–2740 (2003).
Schmitt, C. A. et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 1, 289–298 (2002). An important study in which the contribution of p53 inactivation to lymphomagenesis is assessed in vivo , either in the absence or in the presence of a BCL2 transgene, which actually alleviates the pressure to inactivate p53.
Fridman, J. S. & Lowe, S. W. Control of apoptosis by p53. Oncogene 22, 9030–9040 (2003).
Liu, G. et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nature Genet. 36, 63–68 (2004). A study in which a knock-in mutation of p53, which is supposed to have little or no apoptosis-inducing potential, is examined for its tumour-suppressive effect.
Lawson, B. R. et al. Deficiency of the cyclin kinase inhibitor p21WAF-1/CIP-1 promotes apoptosis of activated/memory T cells and inhibits spontaneous systemic autoimmunity. J. Exp. Med. 199, 547–557 (2004).
Sogame, N., Kim, M. & Abrams, J. M. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl Acad. Sci. USA 100, 4696–4701 (2003).
Cregan, S. P. et al. Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J. Cell Biol. 158, 507–517 (2002).
Alano, C. C., Ying, W. & Swanson, R. A. Poly(ADP-ribose) polymerase-1 mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J. Biol. Chem. 279, 18895–18902 (2004).
Duckett, D. R., Bronstein, S. M., Taya, Y. & Modrich, P. hMutSα- and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl Acad. Sci. USA 96, 12384–12388 (1999).
Hickman, M. J. & Samson, L. D. Apoptotic signaling in response to a single type of DNA lesion, O6-methylguanine. Mol. Cell 14, 105–116 (2004). An exhaustive analysis of the molecular mechanisms that link oxidative DNA damage to activation of the apoptotic default pathway, thereby revealing cross-talk between detection of DNA damage by the DNA-repair machinery and the activation of lethal signals.
Nakayama, H. RecQ family helicases: roles as tumor suppressor proteins. Oncogene 21, 9008–9021 (2002).
Davalos, A. R. & Campisi, J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J. Cell Biol. 162, 1197–1209 (2003).
Cherbonnel-Lasserre, C., Gauny, S. & Kronenberg, A. Suppression of apoptosis by Bcl-2 or Bcl-xL promotes susceptibility to mutagenesis. Oncogene 13, 1489–1497 (1996).
Kuo, M. L., Shiah, S. G., Wang, C. J. & Chuang, S. E. Suppression of apoptosis by Bcl-2 to enhance benzene metabolites-induced oxidative DNA damage and mutagenesis: a possible mechanism of carcinogenesis. Mol. Pharmacol. 55, 894–901 (1999).
Saintigny, Y. et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20, 3861–3870 (2001).
Canitrot, Y. et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood 102, 2632–2637 (2003).
Slupianek, A. et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell 8, 795–806 (2001).
Deutsch, E. et al. BCR–ABL down-regulates the DNA repair protein DNA-PKcs . Blood 97, 2084–2090 (2001).
Skorski, T. BCR/ABL regulates response to DNA damage: the role in resistance to genotoxic treatment and in genomic instability. Oncogene 21, 8591–8604 (2002).
Zhao, R., Yang, F. T. & Alexander, D. R. An oncogenic tyrosine kinase inhibits DNA repair and DNA-damage-induced Bcl-xL deamidation in T cell transformation. Cancer Cell 5, 37–49 (2004). Description of LCK as a kinase that can simultaneously mediate the inhibition of DNA repair, DNA-damage-induced BCL-X L deamidation, BAX conformational change and mitochondrial translocation, cytochrome- c release and the apoptotic caspase execution cascade.
Kerr, J. F. R., Wyllie, A. H. & Currie, A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 (1972).
Green, D. R. & Reed, J. C. Mitochondria and apoptosis. Science 281, 1309–1312 (1998).
Wang, X. The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933 (2002).
Adams, J. M. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17, 2481–2495 (2003).
Zamzami, N. & Kroemer, G. Mitochondria in apoptosis. How Pandora's box opens. Nature Rev. Mol. Cell. Biol. 2, 67–71 (2001).
Igney, F. H. & Krammer, P. H. Death and anti-death: tumour resistance to apoptosis. Nature Rev. Cancer 2, 277–288 (2002).
Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37, 614–636 (1965).
Harley, C. B., Futcher, A. B. & Greider, C. W. Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990).
Yeo, E. J., Hwang, Y. C., Kang, C. M., Choy, H. E. & Park, S. C. Reduction of UV-induced cell death in the human senescent fibroblasts. Mol. Cell 10, 415–422 (2000).
Seluanov, A. et al. Change of the death pathway in senescent human fibroblasts in response to DNA damage is caused by an inability to stabilize p53. Mol. Cell Biol. 21, 1552–1564 (2001).
Schmitt, C. A. et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109, 335–346 (2002). Seminal paper in which premature or inducible senescence, in addition to apoptosis, were identified as a drug-responsive effector programme of cancer therapy. Tumours that have defects in senescence-controlling genes, such as p53 or INK4A , display an impaired treatment response.
Blagosklonny, M. V. Cell senescence and hypermitogenic arrest. EMBO Rep. 4, 358–362 (2003).
Terada, N. et al. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature 416, 542–545 (2002).
Vassilopoulos, G., Wang, P. R. & Russell, D. W. Transplanted bone marrow regenerates liver by cell fusion. Nature 422, 901–904 (2003).
Wang, X. et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature 422, 897–901 (2003). References 94 and 95 show that cell fusion can occur in vivo among normal somatic cells, presumably in physiological conditions.
Goldenberg, D. M., Pavia, R. A. & Tsao, M. C. In vivo hybridisation of human tumour and normal hamster cells. Nature 250, 649–651 (1974).
Fortuna, M. B., Dewey, M. J. & Furmanski, P. Enhanced lung colonization and tumorigenicity of fused cells isolated from primary MCA tumors. Cancer Lett. 55, 109–114 (1990).
Duelli, D. & Lazebnik, Y. Cell fusion: a hidden enemy? Cancer Cell 3, 445–448 (2003). An excellent review that summarizes the evidence on cell fusion as a putative oncogenic event.
Miller, F. R., Mohamed, A. N. & McEachern, D. Production of a more aggressive tumor cell variant by spontaneous fusion of two mouse tumor subpopulations. Cancer Res. 49, 4316–4321 (1989).
Duelli, D. M. & Lazebnik, Y. A. Primary cells suppress oncogene-dependent apoptosis. Nature Cell Biol. 2, 859–862 (2000).
Pawelek, J. M. Tumour cell hybridization and metastasis revisited. Melanoma Res. 10, 507–514 (2000).
Acknowledgements
The authors' own work is supported by the European Commission.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- GENOMIC INSTABILITY
-
The failure to transmit an accurate copy of the entire genome from one cell to its two daughter cells. Note that this term does not describe a state but, rather, a process.
- CENTROSOME
-
A specialized organelle that duplicates during interphase and that constitutes the centre of the mitotic spindle.
- POLYPLOIDY
-
A situation in which a cell has more than two complete sets of chromosomes in G1 or more than four sets in G2/M: so, triploid cells carry 3N in G1 and tetraploid cells have 4N. Note that polyploidy (not to be confused with hyperploidy, a special case of aneuploidy with too many chromosomes) refers to simple multiples of the normal chromosome number.
- CHROMOSOMAL INSTABILITY
-
(CIN). Genetic changes that are manifested at the level of chromatin maintenance and segregation.
- HAPLOINSUFFICIENT
-
A gene that requires bi-allelic expression. Suppression of one allele would reduce the gene dosage below the critical level.
- LINKAGE DISEQUILIBRIUM
-
Non-independent assortment of genes during cell division; for example, because they are situated on the same chromosome.
- MICROSATELLITE INSTABILITY
-
(MSI). Alterations in the length of short repetitive sequences (microsatellites), which can be detected by the PCR amplification of tumour DNA. After PCR, new bands that were not present in PCR products of the corresponding normal DNA will appear if MSI has occurred.
- BCL2 FAMILY
-
A family of proteins that all contain at least one BCL2 homology (BH) region. The family is divided into anti-apoptotic multidomain proteins (such as BCL2 and BCL-XL), which contain four BH domains (BH1, BH2, BH3, BH4), pro-apoptotic multidomain proteins (for example, BAX and BAK), which contain BH1, BH2 and BH3, and the pro-apoptotic BH3-only protein family (for example, BID, BIM and PUMA).
- ANEUPLOIDY
-
The ploidy of a cell refers to the number of chromosome sets that it contains. Aneuploid karyotypes are chromosome complements that are not a simple multiple of the haploid set.
- NON-HOMOLOGOUS END JOINING
-
(NHEJ). The main pathway that is used throughout the cell cycle to repair chromosomal double-strand DNA breaks in somatic cells. In contrast to homologous-recombination repair, NHEJ is error-prone because it leads to the joining of heterologous ends.
- DICENTRIC CHROMOSOME
-
A chromosome that carries two centromeres, which arise from the aberrant fusion of 'naked' telomeres or interstitial double-strand breaks.
- BREAKAGE-FUSION-BRIDGE (BFB) CYCLE
-
Dicentric chromosomes — which are generated by the fusion of two centrosome-containing chromosome fragments or two 'naked' telomeres — break apart in mitosis, after which they fuse again, forming a mutated dicentric chromosome.
- APOPTOSOME
-
A complex that forms when cytochrome c is released from mitochondria and interacts with the cytosolic protein APAF1, which, in turn, recruits pro-caspase-9. In the presence of dATP, this interaction results in the allosteric activation of caspase-9 and in the formation of a caspase-3 activation complex.
- MITOCHONDRIAL-MEMBRANE PERMEABILIZATION
-
(MMP). A pro-apoptotic process whereby mitochondrial membranes undergo transient and stable permeabilization and become structurally reorganized. As a result, proteins that are normally retained in the intermembrane space are released through the outer membrane and the bioenergetic function of mitochondria is compromised.
- CASPASE
-
A family of cysteine proteases that cleave after asparagine residues. Initiator caspases are typically activated in response to particular stimuli (for example, caspase-8 after death-receptor ligation, caspase-9 after apoptosome activation, caspase-2 after DNA damage and PIDD activation), whereas effector caspases (mainly caspase-3, -6 and -7) are particularly important for the ordered dismantling of vital cellular structures.
- SENESCENCE
-
A nearly irreversible stage of permanent G1 cell-cycle arrest, which is linked to morphological changes (flattening of the cells), metabolic changes and changes in gene expression (for example, β-galactosidase). The induction of senescence depends on p53 and cell-cycle inhibitors such as p21 and p16.
- ENDOREPLICATION
-
The replication of DNA during S phase of the cell cycle without the subsequent completion of mitosis.
- PASSENGER PROTEIN
-
A protein that shares a characteristic pattern of association with chromatin in prophase, centromeres in metaphase and early anaphase, and then the midzone and midbody in late anaphase and telophase, respectively.
- ANISOCYTOSIS
-
Abnormal heterogeneity in cell size.
- ANISOKARYOSIS
-
Abnormal heterogeneity in nuclear size and/or in the cytoplasm:nucleus ratio.
- DNA-STRUCTURE CHECKPOINT
-
A checkpoint that arrests cell-cycle advancement until DNA mutations such as double-strand breaks are repaired, or until the replication of complementary strands has been completed.
- CYTOCHROME C
-
A haem protein that is normally confined to the mitochondrial intermembrane space. Following induction of apoptosis, cytochrome c is released from mitochondria and triggers the formation of the apoptosome, a caspase-activation complex.
- APOPTOSIS-INDUCING FACTOR
-
(AIF). A flavoprotein that is normally present in the mitochondrial intermembrane space. Following the induction of apoptosis, AIF translocates to the nucleus where it activates a molecular complex that causes large-scale DNA fragmentation, presumably in a caspase-independent fashion.
- SYNCYTIUM
-
A cytoplasm that contains several nuclei within the same plasma membrane and without internal cell boundaries. Syncytia are mostly generated by cell fusion.
- KARYOKINESIS
-
The physical separation of the daughter nuclei at the end of mitosis.
- SPINDLE-ASSEMBLY CHECKPOINT
-
A checkpoint that monitors the correct attachment of chromosomes to spindles in the metaphase–anaphase transition. Activation of this checkpoint causes cell-cycle arrest as a result of the inhibition of anaphase-promoting complex/cyclosome (APC/C) and is mediated by a cytoplasmic activity known as cytostatic factor (CSF).
- KINETOCHORE
-
A specialized condensed chromosomal region in which the chromatids are held together to form an X shape.
- SMAC/DIABLO
-
A mitochondrial intermembrane protein that, on apoptotic release, can interact with inhibitor of apoptosis (IAP) proteins such as XIAP, thereby inhibiting their function and facilitating caspase activation.
- BASE-EXCISION REPAIR
-
(BER). The main pathway that is responsible for the repair of apurinic and apyrimidinic (AP) sites in DNA. BER is catalyzed in four consecutive steps by a DNA glycosylase, which removes the damaged base; an AP endonuclease, which processes the abasic site; a DNA polymerase, which inserts the new nucleotide(s); and DNA ligase, which rejoins the DNA strand.
- NUCLEOTIDE-EXCISION REPAIR
-
(NER). A process in which a small region of the DNA strand that surrounds DNA damage is removed from the DNA helix as an oligonucleotide.
Rights and permissions
About this article
Cite this article
Zhivotovsky, B., Kroemer, G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol 5, 752–762 (2004). https://doi.org/10.1038/nrm1443
Issue Date:
DOI: https://doi.org/10.1038/nrm1443
This article is cited by
-
FOXM1 is critical for the fitness recovery of chromosomally unstable cells
Cell Death & Disease (2023)
-
CBX4 deletion promotes tumorigenesis under KrasG12D background by inducing genomic instability
Signal Transduction and Targeted Therapy (2023)
-
Ellagic acid effects on testis, sex hormones, oxidative stress, and apoptosis in the relative sterility rat model following busulfan administration
BMC Complementary Medicine and Therapies (2022)
-
Low-temperature extracts of Purple blossoms of basil (Ocimum basilicum L.) intervened mitochondrial translocation contributes prompted apoptosis in human breast cancer cells
Biological Research (2021)
-
AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway
Cell Death & Disease (2020)