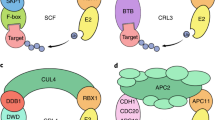
Key Points
-
The ubiquitin-26S proteasome system (UPS) is one of the most complex and pervasive pathways of intracellular protein regulation in plants.
-
Genomic estimates indicate that as much as 6% of the Arabidopsis thaliana transcriptome is devoted to expressing UPS components, with most of these genes encoding ubiquitin ligases (>1,400 loci).
-
Genetic analyses have recently connected the UPS to much of plant hormone signalling, regulation of chromatin structure and transcription, tailoring morphogenesis, responses to environmental challenges, self recognition and the war between pathogens and their plant hosts.
-
In several signalling pathways, components involved in ubiquitin ligation have been shown to act as receptors for hormones and light.
-
Recent proteomic studies have identified a catalogue of plant proteins that are modified by ubiquitylation in planta and have identified various types of polyubiquitin chains.
-
Why the UPS system is so large in plants is currently unclear. Possibilities include their sessile growth habit, which might require additional levels of regulation, the ancient genome duplications common during plant evolution, roles of the UPS in innate immunity and, for annual plants, their reliance on the UPS to confine their life cycle to short growing seasons.
Abstract
Plants, like other eukaryotes, rely on proteolysis to control the abundance of key regulatory proteins and enzymes. Strikingly, genome-wide studies have revealed that the ubiquitin-26S proteasome system (UPS) in particular is an exceedingly large and complex route for protein removal, occupying nearly 6% of the Arabidopsis thaliana proteome. But why is the UPS so pervasive in plants? Data accumulated over the past few years now show that it targets numerous intracellular regulators that have central roles in hormone signalling, the regulation of chromatin structure and transcription, tailoring morphogenesis, responses to environmental challenges, self recognition and battling pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
[No authors listed.] Webster's Collegiate Dictionary (N. E. Merriam-Webster Publishers, Springfield, Massachusetts, 1987).
Wilkinson, K. D., Urban, M. K. & Haas, A. L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 255, 7529–7532 (1980).
Downes, B. & Vierstra, R. D. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem. Soc. Trans. 33, 393–399 (2005).
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell. Dev. Biol. 22, 159–180 (2006).
Smalle, J. & Vierstra, R. D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590 (2004).
Harper, J. W. & Schulman, B. A. Structural complexity in ubiquitin recognition. Cell 124, 1133–1136 (2006).
Mukhopadhyay, D. & Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 (2007).
Dreher, K. & Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99, 787–822 (2007).
Raasi, S. & Wolf, D. H. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin. Cell. Dev. Biol. 18, 780–791 (2007).
Shen, G., Adam, Z. & Zhang, H. The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts. Plant J. 52, 309–321 (2007).
Yan, N., Doelling, J. H., Falbel, T. G., Durski, A. M. & Vierstra, R. D. The ubiquitin-specific protease family from Arabidopsis. At UBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124, 1828–1843 (2000).
Schwechheimer, C. & Calderon Villalobos, L. I. Cullin-containing E3 ubiquitin ligases in plant development. Curr. Opin. Plant Biol. 7, 677–686 (2004).
Gingerich, D. J. et al. Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280, 18810–18821 (2005).
Downes, B. P., Stupar, R. M., Gingerich, D. J. & Vierstra, R. D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35, 729–742 (2003).
Capron, A., Okresz, L. & Genschik, P. First glance at the plant APC/C, a highly conserved ubiquitin–protein ligase. Trends Plant Sci. 8, 83–89 (2003).
Kraft, E. et al. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139, 1597–1611 (2005).
Stone, S. L. et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30 (2005).
Lee, J. H. et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20, 152–167 (2008).
Gagne, J. M., Downes, B. P., Shiu, S.-H., Durski, A. M. & Vierstra, R. D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl Acad. Sci. USA 99, 11519–11524 (2002). This phylogenetic analysis of the A. thaliana F2011 box protein superfamily provided the first indications of the surprising size and complexity of the UPS in plants.
Mudgil, Y., Shiu, S. H., Stone, S. L., Salt, J. N. & Goring, D. R. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134, 59–66 (2004).
Wiborg, J., O'Shea, C. & Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413, 447–457 (2008).
Gingerich, D. J., Hanada, K., Shiu, S. H. & Vierstra, R. D. Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19, 2329–2348 (2007).
Jain, M. et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483 (2007).
Yang, X. et al. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 148, 1189–1200 (2008).
Stone, S. L., Williams, L. A., Farmer, L. M., Vierstra, R. D. & Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428 (2006).
Hotton, S. K. & Callis, J. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59, 467–489 (2008).
Zhang, W. et al. Genetic analysis of CAND1–CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc. Natl Acad. Sci. USA 105, 8470–8475 (2008).
Wei, N. & Deng, X. W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 (2003).
Yang, P. et al. Purification of the Arabidopsis 26S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279, 6401–6413 (2004).
Book, A. J. et al. The RPN5 subunit of the 26S proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21, 460–478 (2009).
Voges, D., Zwickl, P. & Baumeister, W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 (1999).
Husnjak, K. et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008).
Lyapina, S. et al. Promotion of NEDD–CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 (2001).
Fu, H. et al. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 273, 1970–1981 (1998).
Smalle, J. et al. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15, 965–980 (2003).
Smalle, J. et al. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14, 17–32 (2002).
Brukhin, V., Gheyselinck, J., Gagliardini, V., Genschik, P. & Grossniklaus, U. The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17, 2723–2737 (2005).
Ueda, M. et al. The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131, 2101–2111 (2004).
Zhang, F. X. et al. O-GIcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 (2003).
Schmidt, M., Hanna, J., Elsasser, S. & Finley, D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 386, 725–737 (2005).
Kurepa, J., Toh, E. A. & Smalle, J. A. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53, 102–114 (2008).
Ustrell, V., Hoffman, L., Pratt, G. & Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21, 3516–3525 (2002).
Doelling, J. H., Yan, N., Kurepa, J., Walker, J. & Vierstra, R. D. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27, 393–405 (2001).
Hanna, J. et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006).
Doelling, J. H. et al. The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145, 801–813 (2007).
Yang, P. et al. Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J. 51, 441–457 (2007).
Sridhar, V. V. et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447, 735–738 (2007). Found an important connection between histone ubiquitylation and plant epigenetics; it showed that the ubiquitin 2013 histone H2B-specific DUB UBP26 has pleiotropic effects on DNA methylation status and heterochromatin expression.
Luo, M. et al. UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180, 229–236 (2008).
Hatfield, P. M., Gosink, M. M., Carpenter, T. B. & Vierstra, R. D. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 11, 213–226 (1997).
Jin, J. et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 (2004).
Vierstra, R. D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142 (2003).
Schubert, U. et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000).
Bowers, J. E., Chapman, B. A., Rong, J. & Paterson, A. H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 (2003).
Simillion, C., Vandepoele, K., Van Montagu, M. C., Zabeau, M. & Van de Peer, Y. The hidden duplication past of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 99, 13627–13632 (2002).
Clark, R. M. et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317, 338–342 (2007).
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008).
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005).
Dharmasiri, N. et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007). By solving the three-dimensional structure of the TIR1–auxin–domain II complex, this paper showed how the auxin hormone is perceived and how small molecules could regulate the UPS.
Hayashi, K. et al. Small-molecule agonists and antagonists of F-box protein–substrate interactions in auxin perception and signaling. Proc. Natl Acad. Sci. USA 105, 5632–5637 (2008).
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y. & Howe, G. A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. USA 105, 7100–7105 (2008). References 64 and 65 provide compelling evidence that the COI1 F-box protein works similarly to TIR1 as a receptor for a small plant hormone, in this case JA–Ile.
Willige, B. C. et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19, 1209–1220 (2007).
Ueguchi-Tanaka, M. et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155 (2007).
Shimada, A. et al. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523 (2008).
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008). References 68 and 69 describe, with reference to its three-dimensional structure, how the GA receptor GID1 might influence the degradation of Della proteins by SCF complexes containing the SLEEPY 1 and SNEEZY 1 or GID2 F–box proteins.
Joo, S., Liu, Y., Lueth, A. & Zhang, S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140 (2008).
Yoshida, H. et al. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol. Biol. 62, 427–437 (2006).
Chae, H. S., Faure, F. & Kieber, J. J. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15, 545–559 (2003).
Wang, K. L., Yoshida, H., Lurin, C. & Ecker, J. R. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950 (2004).
Christians, M. J. et al. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidosis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345 (2009).
Chen, Y. F. et al. Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J. Biol. Chem. 282, 24752–24758 (2007).
Qiao, H., Chang, K. N., Yazaki, J. & Ecker, J. R. Interplay between ethylene, ETP1/ETP2 F-Box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521 (2009).
Binder, B. M. et al. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19, 509–523 (2007). Provides an example of how multiple E3s could work in concert to temporally regulate a substrate (in this case, key ethylene signalling components) and in doing so generate a tightly regulated and robust response.
Gagne, J. M. et al. Arabidopsis EIN3-binding F-BOX 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl Acad. Sci. USA 101, 6803–6808 (2004).
Guo, H. W. & Ecker, J. R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 (2003).
Potuschak, T. et al. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689 (2003).
Yanagisawa, S., Yoo, S. D. & Sheen, J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525 (2003).
Yoo, S. D., Cho, Y. H., Tena, G., Xiong, Y. & Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795 (2008).
Potushak, T. et al. The exonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18, 3047–3057 (2006).
Konishi, M. & Yanagisawa, S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55, 821–831 (2008).
Olmedo, G. et al. ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl Acad. Sci. USA 103, 13286–13293 (2006).
Stirnberg, P., Furner, I. J. & Ottoline Leyser, H. M. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 (2007).
Gomez-Roldan, V. et al. Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008).
Umehara, M. et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008).
Zhang, X., Garreton, V. & Chua, N. H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543 (2005).
Qin, F. et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707 (2008).
Disch, S. et al. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279 (2006).
Li, Y., Zheng, L., Corke, F., Smith, C. & Bevan, M. W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336 (2008).
Kim, H. J. et al. Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455, 1134–1137 (2008). Describes an example of how spatial expression of an E3 could be used to control the development of specific cell types, in this case the pollen germinative cell that produces the two sperm cells required for double fertilization.
McKim, S. M. et al. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537–1546 (2008).
Hepworth, S. R., Klenz, J. E. & Haughn, G. W. UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223, 769–778 (2006).
Stirnberg, P., van De Sande, K. & Leyser, H. M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141 (2002).
Schwager, K. M. et al. Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 19, 1163–1178 (2007).
Yin, X. J. et al. Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19, 1898–1911 (2007).
Hoecker, U. Regulated proteolysis in light signaling. Curr. Opin. Plant Biol. 8, 469–476 (2005).
Clough, R. C. et al. Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167 (1999).
Yu, X. et al. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19, 3146–3156 (2007).
Al-Sady, B., Ni, W., Kircher, S., Schafer, E. & Quail, P. H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006).
Osterlund, M. T., Hardtke, C. S., Wei, N. & Deng, X. W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000).
Jang, S. et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288 (2008).
Mas, P., Kim, W. Y., Somers, D. E. & Kay, S. A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570 (2003).
Sawa, M., Nusinow, D. A., Kay, S. A. & Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265 (2007). Shows how an E3 could be exploited to measure photoperiod by binding a blue light-absorbing FMN chromophore.
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A. & Kay, S. A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297 (2005).
Kim, W. Y. et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360 (2007).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Azevedo, C. et al. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076 (2002).
Austin, M. J. et al. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080 (2002).
Gonzalez-Lamothe, R. et al. The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083 (2006).
Trujillo, M., Ichimura, K., Casais, C. & Shirasu, K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401 (2008).
Tada, Y. et al. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 (2008).
Janjusevic, R., Abramovitch, R. B., Martin, G. B. & Stebbins, C. E. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226 (2006).
Rosebrock, T. R. et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374 (2007). Shows how pathogens could exploit the UPS during infection by co-opting a domain with ubiquitin ligase activity to help remove a plant sensor of pathogen invasion.
Bortolamiol, D., Pazhouhandeh, M., Marrocco, K., Genschik, P. & Ziegler-Graff, V. The polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621 (2007).
Baumberger, N., Tsai, C. H., Lie, M., Havecker, E. & Baulcombe, D. C. The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614 (2007).
Angot, A. et al. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl Acad. Sci. USA 103, 14620–14625 (2006).
Tzfira, T., Vaidya, M. & Citovsky, V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92 (2004).
Stone, S. L., Anderson, E. M., Mullen, R. T. & Goring, D. R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898 (2003). Provides one of the first connections between the UPS and sporophytic self-incompatibility mechanisms in plants.
Samuel, M. A. et al. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147, 2084–2095 (2008).
Sijacic, P. et al. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429, 302–305 (2004). Provides one of the first connections between the UPS and gametophytic self-incompatibility mechanisms in plants.
Huang, J., Zhao, L., Yang, Q. & Xue, Y. AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-Box protein SLF. Plant J. 46, 780–793 (2006).
Hua, Z. & Kao, T. H. Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18, 2531–2553 (2006).
Hua, Z., Meng, X. & Kao, T. H. Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF like proteins reveals its unique function in S-RNase based self-incompatibility. Plant Cell 19, 3593–3609 (2007).
Weake, V. M. & Workman, J. L. Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 (2008).
Fleury, D. et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19, 417–432 (2007).
Liu, Y., Koornneef, M. & Soppe, W. J. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19, 433–444 (2007).
Kraft, E., Bostick, M., Jacobsen, S. E. & Callis, J. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 56, 704–715 (2008).
Peng, J. et al. A proteomics approach to understanding protein ubiquitination. Nature Biotech. 21, 921–926 (2003).
Kirkpatrick, D. S., Weldon, S. F., Tsaprailis, G., Liebler, D. C. & Gandolfi, A. J. Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 5, 2104–2111 (2005).
Matsumoto, M. et al. Large-scale analysis of the human ubiquitin-related proteome. Proteomics 5, 4145–4151 (2005).
Mayor, T., Graumann, J., Bryan, J., MacCoss, M. J. & Deshaies, R. J. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell. Proteomics 6, 1885–1895 (2007).
Hitchcock, A. L., Auld, K., Gygi, S. P. & Silver, P. A. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl Acad. Sci. USA 100, 12735–12740 (2003).
Maor, R. et al. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell Proteomics 6, 601–610 (2007).
Saracco, S. A. et al. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 9 Mar 2009 (doi: 10.1111/j.1365-313X.2009.03862.x). References 136 and 137 carried out the first proteomic analyses of ubiquitylated proteins in plants. They revealed a number of interesting proteins that are potential targets of the UPS, provided a catalogue of ubiquitin attachment sites and showed that multiple types of ubiquitin polymers are assembled in planta .
Ding, X. et al. A rice kinase-protein interaction map. Plant Physiol. 149, 1478–1492 (2009).
Vijay-Kumar, S. et al. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J. Biol. Chem. 262, 6396–6399 (1987).
Acknowledgements
The author thanks various members of the laboratory for helpful discussions and apologizes to those whose work was not cited because of space constraints. Research in the Vierstra laboratory is supported by grants from the US Department of Energy Basic Energy Sciences Program, the US Department of Agriculture Cooperative State Research, Education and Extension Service, the US National Science Foundation Arabidopsis 2010 Program, and the University of Wisconsin College of Agriculture and Life Sciences.
Author information
Authors and Affiliations
Related links
Glossary
- Ubiquitin fold
-
The three-dimensional protein structure that is characteristic of ubiquitin and related proteins. Its core domain consists of a five-stranded β-sheet that diagonally cradles an α-helix.
- Abscisic acid
-
A plant hormone derived from carotenoids that helps plants respond to stress and prevents precocious germination of seeds.
- RING domain
-
A cross-braced structure formed by an octet of Cys residues and His residues that chelate two zinc atoms. Its threedimensional structure creates a pocket that binds the ubiquitin-E2 intermediate during the ubiquitylation of substrates.
- COP9-signalosome
-
A multi-subunit protein complex that is structurally related to the lid of the 26S proteasome. One subunit removes the ubiquitin fold protein RUB1 from the cullin subunit in CRL E3s.
- Auxin
-
A plant hormone related to indole3-acetic acid (IAA) that regulates plant cell elongation, the activity of the shoot and root meristems and vascular development.
- Jasmonic acid
-
A plant hormone related to oxylipins that is important for plant development and protection from biotic challenges.
- Gibberellin
-
A steroid hormone that promotes cell division and seed germination in plants.
- Ethylene
-
A gaseous plant hormone that inhibits plant growth in response to stress and stimulates fruit ripening.
- Trichome
-
A pointed hair cell (often with three branches) that extends from the surface of aerial portions of plants to provide protection from herbivores and the environment.
Rights and permissions
About this article
Cite this article
Vierstra, R. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10, 385–397 (2009). https://doi.org/10.1038/nrm2688
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2688
This article is cited by
-
The Function of Deubiquitinating Enzymes in Arabidopsis: Recent Progress of Ubiquitin-Specific Proteases (UBPs)
Journal of Plant Biology (2024)
-
The Vitis yeshanensis U-box E3 ubiquitin ligase VyPUB21 enhances resistance to powdery mildew by targeting degradation of NIM1-interacting (NIMIN) protein
Plant Cell Reports (2024)
-
Plant U-box E3 ligases PUB20 and PUB21 negatively regulate pattern-triggered immunity in Arabidopsis
Plant Molecular Biology (2024)
-
Overexpression of a rice Tubby–like protein-encoding gene, OsFBT4, confers tolerance to abiotic stresses
Protoplasma (2023)
-
Role of F-box E3-ubiquitin ligases in plant development and stress responses
Plant Cell Reports (2023)