Key Points
-
Capping protein (CP) is a major regulator of actin assembly dynamics via the capping of actin filament barbed ends. The capping activity of CP can be regulated by a number of different proteins and phospholipids in various ways, some direct and others indirect.
-
The capping protein interacting (CPI) motif is a 30-amino acid region necessary and sufficient to bind and inhibit CP. This motif is found in a set of unrelated proteins, many of which are involved in membrane interactions.
-
CARMIL (capping protein, ARP2/3 and myosin I linker) family proteins contain a CPI motif, and they also contain a separate CARMIL-specific interacting (CSI) motif. In CARMIL, the CPI motif is necessary for distinct cellular functions, such as macropinocytosis.
-
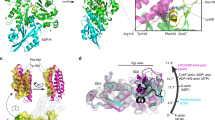
The CPI and CSI motifs are unstructured in the unbound state, but they adopt a specific structure when they bind to CP, applying themselves to the surface of CP. The CPI and CSI motifs decrease the actin capping activity of CP via an allosteric mechanism.
-
The complex of a CPI motif-containing protein with CP retains a low level of capping activity, which raises the possibility that CPI motif-containing proteins may target CP to certain cellular locations, in addition to, or as an alternative to, simply decreasing the capping activity.
-
Vertebrates have three distinct conserved CARMIL genes, which seem to have distinct functions in cells. Of note, CARMIL2 localizes with vimentin filaments, representing a potential novel link between the actin and intermediate filament cytoskeleton systems.
Abstract
Capping protein (CP) binds the fast growing barbed end of the actin filament and regulates actin assembly by blocking the addition and loss of actin subunits. Recent studies provide new insights into how CP and barbed-end capping are regulated. Filament elongation factors, such as formins and ENA/VASP (enabled/vasodilator-stimulated phosphoprotein), indirectly regulate CP by competing with CP for binding to the barbed end, whereas other molecules, including V-1 and phospholipids, directly bind to CP and sterically block its interaction with the filament. In addition, a diverse and unrelated group of proteins interact with CP through a conserved 'capping protein interaction' (CPI) motif. These proteins, including CARMIL (capping protein, ARP2/3 and myosin I linker), CD2AP (CD2-associated protein) and the WASH (WASP and SCAR homologue) complex subunit FAM21, recruit CP to specific subcellular locations and modulate its actin-capping activity via allosteric effects.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
19 September 2014
On page 684 of the above article, there was a mistake in Figure 3b: the PH domain of CARMIL preferentially binds to monophosphorylated membrane lipids rather than to PtdIns(3)P, PtdIns(3,4)P2 or PtdIns(3,4,5)P3. This has been corrected online. We apologize for any confusion caused to readers.
References
Pollard, T. D. & Cooper, J. A. Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009).
Blanchoin, L., Boujemaa-Paterski, R., Sykes, C. & Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014).
Xue, B. & Robinson, R. C. Guardians of the actin monomer. Eur. J. Cell Biol. 92, 316–332 (2013).
Suetsugu, S. & Gautreau, A. Synergistic BAR–NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 22, 141–150 (2012).
Hall, A. Rho family GTPases. Biochem. Soc. Trans. 40, 1378–1382 (2012).
Dominguez, R. Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 20, 217–225 (2010).
Chesarone, M., Gould, C. J., Moseley, J. B. & Goode, B. L. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell 16, 292–302 (2009).
Dominguez, R. & Namgoong, S. in Comprehensive Biophysics (ed. Egelman, E. H. ) 31–47 (Elsevier, 2012).
Bernstein, B. W. & Bamburg, J. R. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195 (2010).
Poukkula, M., Kremneva, E., Serlachius, M. & Lappalainen, P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton 68, 471–490 (2011).
Cooper, J. A. & Sept, D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell. Mol. Biol. 267, 183–206 (2008).
Mejillano, M. R. et al. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118, 363–373 (2004). Shows that loss of CP in migrating cultured cells leads to loss of ARP2/3-based lamellipodia and increased numbers of filopodia.
Isenberg, G., Aebi, U. & Pollard, T. D. An actin-binding protein from Acanthamoeba regulates actin filament polymerization and interactions. Nature 288, 455–459 (1980). Discovers CP by biochemical purification from A. castellanii.
Casella, J. F., Craig, S. W., Maack, D. J. & Brown, A. E. Cap Z(36/32), a barbed end actin-capping protein, is a component of the Z-line of skeletal muscle. J. Cell Biol. 105, 371–379 (1987).
Maruyama, K. et al. β-actinin is equivalent to Cap Z protein. J. Biol. Chem. 265, 8712–8715 (1990).
Schleicher, M., Gerisch, G. & Isenberg, G. New actin-binding proteins from Dictyostelium discoideum. EMBO J. 3, 2095–2100 (1984).
Nag, S., Larsson, M., Robinson, R. C. & Burtnick, L. D. Gelsolin: the tail of a molecular gymnast. Cytoskeleton 70, 360–384 (2013).
Li, G. H., Arora, P. D., Chen, Y., McCulloch, C. A. & Liu, P. Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 32, 999–1025 (2012).
Fowler, V. M. The human erythrocyte plasma membrane: a Rosetta Stone for decoding membrane-cytoskeleton structure. Curr. Top. Membr. 72, 39–88 (2013).
Higgs, H. N. There goes the neighbourhood: Eps8 joins the barbed-end crowd. Nature Cell Biol. 6, 1147–1149 (2004).
Bisi, S. et al. Membrane and actin dynamics interplay at lamellipodia leading edge. Curr. Opin. Cell Biol. 25, 565–573 (2013).
Bruck, S. et al. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J. Biol. Chem. 281, 19196–19203 (2006). Identifies the CPI motif by analysis of CD2AP.
Hernandez-Valladares, M. et al. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nature Struct. Mol. Biol. 17, 497–503 (2010). The structural and biochemical analysis of complexes of CPI and CSI motifs with CP.
Hart, M. C. & Cooper, J. A. Vertebrate isoforms of actin capping protein β have distinct functions In vivo. J. Cell Biol. 147, 1287–1298 (1999).
Schafer, D. A. Inhibition of CapZ during myofibrillogenesis alters assembly of actin filaments. J. Cell Biol. 128, 61–70 (1995).
Schafer, D. A., Korshunova, Y. O., Schroer, T. A. & Cooper, J. A. Differential localization and sequence analysis of capping protein β-subunit isoforms of vertebrates. J. Cell Biol. 127, 453–465 (1994).
Ydenberg, C. A., Smith, B. A., Breitsprecher, D., Gelles, J. & Goode, B. L. Cease-fire at the leading edge: new perspectives on actin filament branching, debranching, and cross-linking. Cytoskeleton 68, 596–602 (2011).
Miyoshi, T. et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175, 947–955 (2006).
Iwasa, J. H. & Mullins, R. D. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17, 395–406 (2007).
Akin, O. & Mullins, R. D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851 (2008).
Pollard, T. D. & Borisy, G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 (2003).
Schaub, S., Meister, J. J. & Verkhovsky, A. B. Analysis of actin filament network organization in lamellipodia by comparing experimental and simulated images. J. Cell Sci. 120, 1491–1500 (2007).
Vinzenz, M. et al. Actin branching in the initiation and maintenance of lamellipodia. J. Cell Sci. 125, 2775–2785 (2012).
Bugyi, B. & Carlier, M. F. Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39, 449–470 (2010).
Lai, F. P. et al. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 (2008).
Yamashita, A., Maeda, K. & Maeda, Y. Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 22, 1529–1538 (2003). Reveals the crystal structure of CP and proposes a model for barbed-end capping.
Sizonenko, G. I., Karpova, T. S., Gattermeir, D. J. & Cooper, J. A. Mutational analysis of capping protein function in Saccharomyces cerevisiae. Mol. Biol. Cell 7, 1–15 (1996).
Schafer, D. A., Jennings, P. B. & Cooper, J. A. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996). Reports dynamic uncapping of CP by polyphosphoinositides.
Kim, T., Cooper, J. A. & Sept, D. The interaction of capping protein with the barbed end of the actin filament. J. Mol. Biol. 404, 794–802 (2010).
Wear, M. A., Yamashita, A., Kim, K., Maeda, Y. & Cooper, J. A. How capping protein binds the barbed end of the actin filament. Curr. Biol. 13, 1531–1537 (2003).
Narita, A. & Maeda, Y. Molecular determination by electron microscopy of the actin filament end structure. J. Mol. Biol. 365, 480–501 (2007).
Narita, A., Takeda, S., Yamashita, A. & Maeda, Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 25, 5626–5633 (2006).
Breitsprecher, D. & Goode, B. L. Formins at a glance. J. Cell Sci. 126, 1–7 (2013).
Zigmond, S. H. et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 (2003).
Ramalingam, N. et al. Phospholipids regulate localization and activity of mDia1 formin. Eur. J. Cell Biol. 89, 723–732 (2010).
Gorelik, R., Yang, C., Kameswaran, V., Dominguez, R. & Svitkina, T. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol. Biol. Cell 22, 189–201 (2011).
Watanabe, N. et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16, 3044–3056 (1997).
Evangelista, M. et al. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122 (1997).
Li, F. & Higgs, H. N. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003).
Chesarone, M. A., Dupage, A. G. & Goode, B. L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nature Rev. Mol. Cell Biol. 11, 62–74 (2010).
Bartolini, F., Ramalingam, N. & Gundersen, G. G. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol. Biol. Cell 23, 4032–4040 (2012).
Paul, A. S. & Pollard, T. D. Review of the mechanism of processive actin filament elongation by formins. Cell. Motil. Cytoskeleton 66, 606–617 (2009).
Goode, B. L. & Eck, M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 (2007).
Drees, F. & Gertler, F. B. Ena/VASP: proteins at the tip of the nervous system. Curr. Opin. Neurobiol. 18, 53–59 (2008).
Walders-Harbeck, B., Khaitlina, S. Y., Hinssen, H., Jockusch, B. M. & Illenberger, S. The vasodilator-stimulated phosphoprotein promotes actin polymerisation through direct binding to monomeric actin. FEBS Lett. 529, 275–280 (2002).
Chereau, D. & Dominguez, R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J. Struct. Biol. 155, 195–201 (2006).
Bear, J. E. et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521 (2002).
Barzik, M. et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280, 28653–28662 (2005). Shows that ENA/VASP proteins promote elongation in the presence of CP.
Breitsprecher, D. et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943–2954 (2008).
Breitsprecher, D. et al. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30, 456–467 (2011).
Hansen, S. D. & Mullins, R. D. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 (2010).
Winkelman, J. D., Bilancia, C. G., Peifer, M. & Kovar, D. R. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl Acad. Sci. USA 111, 4121–4126 (2014).
Mahoney, N. M., Janmey, P. A. & Almo, S. C. Structure of the profilin–poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nature Struct. Biol. 4, 953–960 (1997).
Ferron, F., Rebowski, G., Lee, S. H. & Dominguez, R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 26, 4597–4606 (2007).
Pasic, L., Kotova, T. & Schafer, D. A. Ena/VASP proteins capture actin filament barbed ends. J. Biol. Chem. 283, 9814–9819 (2008).
Yang, Y., Nanduri, S., Sen, S. & Qin, J. The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin. Structure 6, 619–626 (1998).
Taoka, M. et al. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J. Biol. Chem. 278, 5864–5870 (2003). Reports that V-1 interacts with CP.
Taoka, M. et al. Murine cerebellar neurons express a novel gene encoding a protein related to cell cycle control and cell fate determination proteins. J. Biol. Chem. 269, 9946–9951 (1994).
Sen, S. et al. Myotrophin: purification of a novel peptide from spontaneously hypertensive rat heart that influences myocardial growth. J. Biol. Chem. 265, 16635–16643 (1990).
Sil, P., Misono, K. & Sen, S. Myotrophin in human cardiomyopathic heart. Circ. Res. 73, 98–108 (1993).
Sil, P., Mukherjee, D. & Sen, S. Quantification of myotrophin from spontaneously hypertensive and normal rat hearts. Circ. Res. 76, 1020–1027 (1995).
Gupta, S., Purcell, N. H., Lin, A. & Sen, S. Activation of nuclear factor-κB is necessary for myotrophin-induced cardiac hypertrophy. J. Cell Biol. 159, 1019–1028 (2002).
Das, B. et al. Nuclear co-translocation of myotrophin and p65 stimulates myocyte growth. Regulation by myotrophin hairpin loops. J. Biol. Chem. 283, 27947–27956 (2008).
Gupta, S., Maitra, R., Young, D., Gupta, A. & Sen, S. Silencing the myotrophin gene by RNA interference leads to the regression of cardiac hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 297, H627–H636 (2009).
Bhattacharya, N., Ghosh, S., Sept, D. & Cooper, J. A. Binding of myotrophin/V-1 to actin-capping protein: implications for how capping protein binds to the filament barbed end. J. Biol. Chem. 281, 31021–31030 (2006).
Zwolak, A., Fujiwara, I., Hammer, J. A. & Tjandra, N. Structural basis for capping protein sequestration by myotrophin (V-1). J. Biol. Chem. 285, 25767–25781 (2010). Reveals the mechanism of V-1 interaction with CP.
Takeda, S. et al. Two distinct mechanisms for actin capping protein regulation — steric and allosteric inhibition. PLoS Biol. 8, e1000416 (2010). Identified contrasting steric and allosteric mechanisms to regulate CP.
Yin, H. L. & Janmey, P. A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65, 761–789 (2003).
Saarikangas, J., Zhao, H. & Lappalainen, P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289 (2010).
Heiss, S. G. & Cooper, J. A. Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry 30, 8753–8758 (1991).
Kim, K. et al. Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J. Biol. Chem. 282, 5871–5879 (2007).
Kuhn, J. R. & Pollard, T. D. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282, 28014–28024 (2007).
Li, J. et al. Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24, 3742–3754 (2012).
Xu, P., Zot, A. S. & Zot, H. G. Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J. Biol. Chem. 270, 25316–25319 (1995). Discovers Acan125, the first CARMIL, in A. castellanii.
Jung, G., Remmert, K., Wu, X., Volosky, J. M. & Hammer, J. A. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479–1497 (2001). Characterizes D. discoideum CARMIL interactions and mutant phenotypes.
Yang, C. et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 9, 209–221 (2005). Shows that mammalian CARMIL inhibits CP and causes uncapping. Also shows that the CARMIL1–CP complex retains capping activity, which suggests an allosteric mechanism.
Liang, Y., Niederstrasser, H., Edwards, M., Jackson, C. E. & Cooper, J. A. Distinct roles for CARMIL isoforms in cell migration. Mol. Biol. Cell 20, 5290–5305 (2009). Reports that CARMIL1 and CARMIL2 have distinct functions in vertebrate cells.
Fujiwara, I., Remmert, K., Piszczek, G. & Hammer, J. A. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc. Natl Acad. Sci. USA 111, E1970–E1979 (2014).
Edwards, M., Liang, Y., Kim, T. & Cooper, J. A. Physiological role of the interaction between CARMIL1 and capping protein. Mol. Biol. Cell 24, 3047–3055 (2013). Shows the physiological relevance of the CARMIL1–CP interaction.
Brzeska, H., Guag, J., Remmert, K., Chacko, S. & Korn, E. D. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285, 5738–5747 (2010).
Helfand, B. T. et al. Vimentin organization modulates the formation of lamellipodia. Mol. Biol. Cell 22, 1274–1289 (2011).
Cote, J. F. & Vuori, K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17, 383–393 (2007).
Hsu, C. C. et al. Identifying LRRC16B as an oncofetal gene with transforming enhancing capability using a combined bioinformatics and experimental approach. Oncogene 30, 654–667 (2011).
Zwolak, A. et al. CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nature Commun. 4, 2523 (2013). Determines the structure of a large N-terminal fragment of CARMIL, and identifies and characterizes the PH and HD domains.
Kobe, B. & Kajava, A. V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732 (2001).
Bella, J., Hindle, K. L., McEwan, P. A. & Lovell, S. C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 65, 2307–2333 (2008).
Lee, W. L., Ostap, E. M., Zot, H. G. & Pollard, T. D. Organization and ligand binding properties of the tail of Acanthamoeba myosin-IA. Identification of an actin-binding site in the basic (tail homology-1) domain. J. Biol. Chem. 274, 35159–35171 (1999).
Zot, H. G., Bhaskara, V. & Liu, L. Acan125 binding to the SH3 domain of acanthamoeba myosin-IC. Arch. Biochem. Biophys. 375, 161–164 (2000).
Bosc, D. G. et al. Identification and characterization of CKIP-1, a novel pleckstrin homology domain-containing protein that interacts with protein kinase CK2. J. Biol. Chem. 275, 14295–14306 (2000).
Canton, D. A. et al. The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol. Cell. Biol. 25, 3519–3534 (2005).
Canton, D. A., Olsten, M. E., Niederstrasser, H., Cooper, J. A. & Litchfield, D. W. The role of CKIP-1 in cell morphology depends on its interaction with actin-capping protein. J. Biol. Chem. 281, 36347–36359 (2006). Reports that the interaction of CKIP1 with CP is physiologically relevant in cells.
Nie, J. et al. CKIP-1: a scaffold protein and potential therapeutic target integrating multiple signaling pathways and physiological functions. Ageing Res. Rev. 12, 276–281 (2013).
Olsten, M. E., Canton, D. A., Zhang, C., Walton, P. A. & Litchfield, D. W. The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J. Biol. Chem. 279, 42114–42127 (2004).
Zhang, L. et al. Role for the pleckstrin homology domain-containing protein CKIP-1 in AP-1 regulation and apoptosis. EMBO J. 24, 766–778 (2005).
Tokuda, E. et al. Casein kinase 2-interacting protein-1, a novel Akt pleckstrin homology domain-interacting protein, down-regulates PI3K/Akt signaling and suppresses tumor growth in vivo. Cancer Res. 67, 9666–9676 (2007).
Zhang, L. et al. CKIP-1 recruits nuclear ATM partially to the plasma membrane through interaction with ATM. Cell. Signal. 18, 1386–1395 (2006).
Zhang, L. et al. The PH domain containing protein CKIP-1 binds to IFP35 and Nmi and is involved in cytokine signaling. Cell. Signal. 19, 932–944 (2007).
Lu, K. et al. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nature Cell Biol. 10, 994–1002 (2008).
Nie, J. et al. CKIP-1 acts as a colonic tumor suppressor by repressing oncogenic Smurf1 synthesis and promoting Smurf1 autodegradation. Oncogene 33, 3677–3687 (2013).
Derivery, E. et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 (2009).
Duleh, S. N. & Welch, M. D. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton 67, 193–206 (2010).
Gomez, T. S. & Billadeau, D. D. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 (2009).
Gomez, T. S., Gorman, J. A., de Narvajas, A. A., Koenig, A. O. & Billadeau, D. D. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215–3228 (2012).
Zech, T. et al. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124, 3753–3759 (2011).
Jia, D. et al. WASH and WAVE actin regulators of the Wiskott–Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl Acad. Sci. USA 107, 10442–10447 (2010). Shows that FAM21 interacts with CP in the WASH complex.
Harbour, M. E., Breusegem, S. Y. & Seaman, M. N. Recruitment of the endosomal WASH complex is mediated by the extended 'tail' of Fam21 binding to the retromer protein Vps35. Biochem. J. 442, 209–220 (2012).
Jia, D., Gomez, T. S., Billadeau, D. D. & Rosen, M. K. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361 (2012).
Park, L. et al. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev. Cell 24, 169–181 (2013). Identifies the trafficking function of FAM21 and CP in the WASH complex.
Dustin, M. L. et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 94, 667–677 (1998).
Shih, N. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science 286, 312–315 (1999).
Yaddanapudi, S. et al. CD2AP in mouse and human podocytes controls a proteolytic program that regulates cytoskeletal structure and cellular survival. J. Clin. Invest. 121, 3965–3980 (2011).
Kirsch, K. H., Georgescu, M.-M., Ishimaru, S. & Hanafusa, H. CMS: An adapter molecule involved in cytoskeletal rearrangements. Proc. Natl Acad. Sci. USA 96, 6211–6216 (1999).
Gaidos, G., Soni, S., Oswald, D. J., Toselli, P. A. & Kirsch, K. H. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J. Cell Sci. 120, 2366–2377 (2007).
Kirsch, K. H. et al. The adapter type protein CMS/CD2AP binds to the proto-oncogenic protein c-Cbl through a tyrosine phosphorylation-regulated Src homology 3 domain interaction. J. Biol. Chem. 276, 4957–4963 (2001).
Roldan, J. L., Blackledge, M., van Nuland, N. A. & Azuaga, A. I. Solution structure, dynamics and thermodynamics of the three SH3 domains of CD2AP. J. Biomol. NMR 50, 103–117 (2011).
Zhao, J. et al. CD2AP links cortactin and capping protein at the cell periphery to facilitate formation of lamellipodia. Mol. Cell. Biol. 33, 38–47 (2013). Reports that CD2AP recruits CP and cortactin to the plasma membrane.
Eyers, C. E. et al. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 389, 127–135 (2005). Shows that phosphorylation of CapZIP inhibits its interaction with CP.
Palmgren, S., Vartiainen, M. & Lappalainen, P. Twinfilin, a molecular mailman for actin monomers. J. Cell Sci. 115, 881–886 (2002).
Helfer, E. et al. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 25, 1184–1195 (2006).
Palmgren, S., Ojala, P. J., Wear, M. A., Cooper, J. A. & Lappalainen, P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155, 251–260 (2001).
Vartiainen, M. K., Sarkkinen, E. M., Matilainen, T., Salminen, M. & Lappalainen, P. Mammals have two twinfilin isoforms whose subcellular localizations and tissue distributions are differentially regulated. J. Biol. Chem. 278, 34347–34355 (2003).
Falck, S. et al. Biological role and structural mechanism of twinfilin-capping protein interaction. EMBO J. 23, 3010–3019 (2004).
Nevalainen, E. M., Skwarek-Maruszewska, A., Braun, A., Moser, M. & Lappalainen, P. Two biochemically distinct and tissue-specific twinfilin isoforms are generated from the mouse Twf2 gene by alternative promoter usage. Biochem. J. 417, 593–600 (2009).
Ojala, P. J. et al. The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell 13, 3811–3821 (2002).
Goode, B. L., Drubin, D. G. & Lappalainen, P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J. Cell Biol. 142, 723–733 (1998).
Wahlstrom, G. et al. Twinfilin is required for actin-dependent developmental processes in Drosophila. J. Cell Biol. 155, 787–796 (2001).
Uruno, T., Remmert, K. & Hammer, J. A. CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J. Biol. Chem. 281, 10635–10650 (2006). Shows that CARMIL inhibits CP and uncaps actin filaments, and that full-length CARMIL is autoinhibited.
Zwolak, A., Uruno, T., Piszczek, G., Hammer, J. A. & Tjandra, N. Molecular basis for barbed end uncapping by CARMIL homology domain 3 of mouse CARMIL-1. J. Biol. Chem. 285, 29014–29026 (2010).
Kim, T., Ravilious, G. E., Sept, D. & Cooper, J. A. Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J. Biol. Chem. 287, 15251–15262 (2012). Reports the allosteric mechanism for CARMIL-mediated inhibition of CP.
Takeda, S. et al. Actin capping protein and its inhibitor CARMIL: how intrinsically disordered regions function. Phys. Biol. 8, 035005 (2011).
Fujiwara, I., Remmert, K. & Hammer, J. A. Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J. Biol. Chem. 285, 2707–2720 (2010). Uses single-molecule imaging to show uncapping by CARMIL.
Abbasi, A. A. Piecemeal or big bangs: correlating the vertebrate evolution with proposed models of gene expansion events. Nature Rev. Genet. 11, 166 (2010).
Sinnar, S. A., Antoku, S., Saffin, J. M., Cooper, J. A. & Halpain, S. Capping protein is essential for cell migration in vivo and for filopodial morphology and dynamics. Mol. Biol. Cell 25, 2152–2160 (2014).
Svitkina, T. M. et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421 (2003).
Avenarius, M. R. et al. Correlation of actin crosslinker and capper expression levels with stereocilia growth phases. Mol. Cell Proteom. 13, 606–620 (2014).
Tang, V. W. & Brieher, W. M. FSGS3/CD2AP is a barbed-end capping protein that stabilizes actin and strengthens adherens junctions. J. Cell Biol. 203, 815–833 (2013).
Cassimeris, L., Safer, D., Nachmias, V. T. & Zigmond, S. H. Thymosin β4 sequesters the majority of G-actin in resting human polymorphonuclear leukoctyes. J. Cell Biol. 119, 1261–1270 (1992).
Remmert, K. et al. CARMIL is a bona fide capping protein interactant. J. Biol. Chem. 279, 3068–3077 (2004). Shows that CARMIL purified from cells contains CP and that it exists as a dimer.
Cao, L. G. Mechanism of the formation of contractile ring in dividing cultured animal cells. I. Recruitment of preexisting actin filaments into the cleavage furrow. J. Cell Biol. 110, 1089–1095 (1990).
Liang, Y. et al. The lymphoid lineage-specific actin-uncapping protein Rltpr is essential for costimulation via CD28 and the development of regulatory T cells. Nature Immunol. 14, 858–866 (2013).
Shih, N. Y. et al. CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. Am. J. Pathol. 159, 2303–2308 (2001).
Eichinger, L., Noegel, A. A. & Schleicher, M. Domain structure in actin-binding proteins: expression and functional characterization of truncated severin. J. Cell Biol. 112, 665–676 (1991).
Lehtonen, S. et al. In vivo interaction of the adapter protein CD2-associated protein with the type 2 polycystic kidney disease protein, polycystin-2. J. Biol. Chem. 275, 32888–32893 (2000).
Drenckhahn, D. et al. Three different actin filament assemblies occur in every hair cell: each contains a specific actin crosslinking protein. J. Cell Biol. 112, 641–651 (1991).
Cormont, M. et al. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic 4, 97–112 (2003).
Take, H. et al. Cloning and characterization of a novel adaptor protein, CIN85, that interacts with c-Cbl. Biochem. Biophys. Res. Commun. 268, 321–328 (2000).
Gout, I. et al. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J. 19, 4015–4025 (2000).
Lynch, D. K. et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278, 21805–21813 (2003).
Schwarz, K. et al. Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J. Clin. Invest. 108, 1621–1629 (2001).
Brett, T. J., Traub, L. M. & Fremont, D. H. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10, 797–809 (2002).
Huber, T. B. et al. Nephrin and CD2AP associate with phosphoinositide 3-OH kinase and stimulate AKT-dependent signaling. Mol. Cell. Biol. 23, 4917–4928 (2003).
Asanuma, K., Campbell, K. N., Kim, K., Faul, C. & Mundel, P. Nuclear relocation of the nephrin and CD2AP-binding protein dendrin promotes apoptosis of podocytes. Proc. Natl Acad. Sci. USA 104, 10134–10139 (2007).
Kowanetz, K. et al. CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol. Biol. Cell 15, 3155–3166 (2004).
Liu, Y., Yerushalmi, G. M., Grigera, P. R. & Parsons, J. T. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 280, 8884–8892 (2005).
Dominguez, R. Actin-binding proteins — a unifying hypothesis. Trends Biochem. Sci. 29, 572–578 (2004).
Frost, A., De Camilli, P. & Unger, V. M. F-BAR proteins join the BAR family fold. Structure 15, 751–753 (2007).
Safi, A. et al. Role for the pleckstrin homology domain-containing protein CKIP-1 in phosphatidylinositol 3-kinase-regulated muscle differentiation. Mol. Cell. Biol. 24, 1245–1255 (2004).
Baas, D. et al. CKIP-1 regulates mammalian and zebrafish myoblast fusion. J. Cell Sci. 125, 3790–3800 (2012).
Franck, Z. Microinjection of villin into cultured cells induces rapid and long- lasting changes in cell morphology but does not inhibit cytokinesis, cell motility, or membrane ruffling. J. Cell Biol. 111, 2475–2485 (1990).
Sakamoto, T. et al. CKIP-1 is an intrinsic negative regulator of T-cell activation through an interaction with CARMA1. PLoS ONE 9, e85762 (2014).
Ling, S. et al. CKIP-1 inhibits cardiac hypertrophy by regulating class II histone deacetylase phosphorylation through recruiting PP2A. Circulation 126, 3028–3040 (2012).
van Duijn, T. J., Anthony, E. C., Hensbergen, P. J., Deelder, A. M. & Hordijk, P. L. Rac1 recruits the adapter protein CMS/CD2AP to cell-cell contacts. J. Biol. Chem. 285, 20137–20146 (2010).
Fukui, Y. Structure and function of the cytoskeleton of a Dictyostelium myosin- defective mutant. J. Cell Biol. 110, 367–378 (1990).
de Arruda, M. V., Watson, S., Lin, C. S., Leavitt, J. & Matsudaira, P. Fimbrin is a homologue of the cytoplasmic phosphoprotein plastin and has domains homologous with calmodulin and actin gelation proteins. J. Cell Biol. 111, 1069–1079 (1990).
Helfer, E. et al. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol. Cell 105, 191–207 (2013).
Acknowledgements
The authors thank members of their laboratories for comments on the manuscript and contributions to the work discussed in this article. They also thank R. Cheney and R. Insall for comments on the manuscript. Work in the authors' laboratories and the writing of this manuscript was supported by National Institutes of Health (NIH) grants GM038542 and GM095509 (to J.A.C.) and GM073791 and MH087950 (to R.D.). M.E. was supported by NIH training grant 5T90DA02287104.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Sarcomere Z-disks
-
The sarcomere is the basic unit of the contractile apparatus of striated muscle. The Z-disk is the structure at each end of the sarcomere to which the barbed ends of the actin-based thin filaments are anchored.
- BAR-domain proteins
-
BAR (Bin–amphiphysin–Rvs) domains of proteins bind to membranes, sensing and inducing curvature. They have roles in membrane trafficking.
- Thin filaments
-
The sarcomere contains thin filaments composed of actin and thick filaments composed of myosin. Their interaction and sliding causes contraction of the sarcomere and hence the muscle.
- Cortical actin
-
Actin filaments that are located close to the plasma membrane. These filaments control the shape and movement of the plasma membrane, and they are often highly dynamic.
- Macropinocytosis
-
Engulfment of extracellular fluid by ruffling and folding back of the plasma membrane. Highly dependent on dynamic actin assembly.
- Vimentin
-
A protein that forms intermediate filaments in cells. Found in a wide range of cell types.
- Retromer
-
A protein complex of endosomes that is involved in membrane receptor recycling.
Rights and permissions
About this article
Cite this article
Edwards, M., Zwolak, A., Schafer, D. et al. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol 15, 677–689 (2014). https://doi.org/10.1038/nrm3869
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3869
This article is cited by
-
Disulfidptosis-associated lncRNAs predict breast cancer subtypes
Scientific Reports (2023)
-
The multiple links between actin and mitochondria
Nature Reviews Molecular Cell Biology (2023)
-
Whole-genome screens reveal regulators of differentiation state and context-dependent migration in human neutrophils
Nature Communications (2023)
-
Multicomponent regulation of actin barbed end assembly by twinfilin, formin and capping protein
Nature Communications (2023)
-
Maternal methionine supplementation during gestation alters alternative splicing and DNA methylation in bovine skeletal muscle
BMC Genomics (2021)