Key Points
-
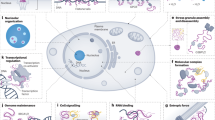
Intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) of proteins that may also contain structured domains mediate crucial signalling processes in eukaryotic cells.
-
Disorder is advantageous in cell signalling because disordered sequences have the potential to bind to multiple partners, often using different structures.
-
Disordered regions are relatively accessible, often contain multiple binding motifs and are frequently the sites for post-translational modification, an important mediator of the control of signalling pathways.
-
Disordered proteins have central roles in the formation of higher-order signalling assemblies and in the operation of circadian clocks.
Abstract
Intrinsically disordered proteins (IDPs) are important components of the cellular signalling machinery, allowing the same polypeptide to undertake different interactions with different consequences. IDPs are subject to combinatorial post-translational modifications and alternative splicing, adding complexity to regulatory networks and providing a mechanism for tissue-specific signalling. These proteins participate in the assembly of signalling complexes and in the dynamic self-assembly of membrane-less nuclear and cytoplasmic organelles. Experimental, computational and bioinformatic analyses combine to identify and characterize disordered regions of proteins, leading to a greater appreciation of their widespread roles in biological processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wright, P. E. & Dyson, H. J. Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J. Mol. Biol. 293, 321–331 (1999).
Dunker, A. K., Brown, C. J., Lawson, J. D., Iakoucheva, L. M. & Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 41, 6573–6582 (2002).
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nature Rev. Mol. Cell Biol. 6, 197–208 (2005).
van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014).
Dunker, A. K., Cortese, M. S., Romero, P., Iakoucheva, L. M. & Uversky, V. N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272, 5129–5148 (2005).
Kim, P. M., Sboner, A., Xia, Y. & Gerstein, M. The role of disorder in interaction networks: a structural analysis. Mol. Systems Biol. 4, 179 (2008).
Iakoucheva, L. M., Brown, C. J., Lawson, J. D., Obradovic, Z. & Dunker, A. K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323, 573–584 (2002).
Liu, J. et al. Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888 (2006).
Galea, C. A., Wang, Y., Sivakolundu, S. G. & Kriwacki, R. W. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry 47, 7598–7609 (2008).
Gsponer, J., Futschik, M. E., Teichmann, S. A. & Babu, M. M. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322, 1365–1368 (2008).
Vavouri, T., Semple, J. I., Garcia-Verdugo, R. & Lehner, B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138, 198–208 (2009).
Babu, M. M., van der Lee, R., de Groot, N. S. & Gsponer, J. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 21, 432–440 (2011).
Tompa, P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354 (2005).
Frey, S., Richter, R. P. & Görlich, D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006).
Tompa, P. Structure and Function of Intrinsically Disordered Proteins (Chapman & Hall/CRC, 2009).
Guharoy, M., Szabo, B., Martos, S. C., Kosol, S. & Tompa, P. Intrinsic structural disorder in cytoskeletal proteins. Cytoskeleton 70, 550–571 (2013).
Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012).
Weber, S. C. & Brangwynne, C. P. Getting RNA and protein in phase. Cell 149, 1188–1191 (2012).
Oldfield, C. J. et al. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry 44, 12454–12470 (2005).
Pontius, B. W. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci. 18, 181–186 (1993).
Stein, A., Pache, R. A., Bernadó, P., Pons, M. & Aloy, P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 276, 5390–5405 (2009).
Gsponer, J. & Babu, M. M. The rules of disorder or why disorder rules. Progress Biophys. Mol. Biol. 99, 94–103 (2009).
Lee, C. W., Ferreon, J. C., Ferreon, A. C., Arai, M. & Wright, P. E. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl Acad. Sci. USA 107, 19290–19295 (2010).
Borg, M. et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc. Natl Acad. Sci. USA 104, 9650–9655 (2007).
Van Roey, K., Dinkel, H., Weatheritt, R. J., Gibson, T. J. & Davey, N. E. The switches. ELM resource: a compendium of conditional regulatory interaction interfaces. Sci. Signal. 6, rs7 (2013).
Van Roey, K., Gibson, T. J. & Davey, N. E. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 22, 378–385 (2012).
Oates, M. E. et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 41, D508–D516 (2013).
Marsh, J. A. & Forman-Kay, J. D. Ensemble modeling of protein disordered states: experimental restraint contributions and validation. Proteins 80, 556–572 (2012).
Salmon, L. et al. NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 132, 8407–8418 (2010).
Sibille, N. & Bernadó, P. Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochem. Soc. Trans. 40, 955–962 (2012).
Varadi, M. et al. pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res. 42, D326–D335 (2014).
Ferreon, A. C., Moran, C. R., Gambin, Y. & Deniz, A. A. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 472, 179–204 (2010).
Hofmann, H. et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl Acad. Sci. USA 109, 16155–16160 (2012).
Das, R. K. & Pappu, R. V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl Acad. Sci. USA 110, 13392–13397 (2013).
Dyson, H. J. & Wright, P. E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12, 54–60 (2002).
Mohan, A. et al. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 (2006).
Neduva, V. & Russell, R. B. Linear motifs: Evolutionary interaction switches. FEBS Lett. 579, 3342–3345 (2005).
Tompa, P., Davey, N. E., Gibson, T. J. & Babu, M. M. A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 (2014).
Demarest, S. J. et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415, 549–553 (2002).
Waters, L. et al. Structural diversity in p160/CREB-binding protein coactivator complexes. J. Biol. Chem. 281, 14787–14795 (2006).
Qin, B. Y. et al. Crystal Structure of IRF-3 in complex with CBP. Structure 13, 1269–1277 (2005).
Spolar, R. S. & Record, M. T. Coupling of local folding to site-specific binding of proteins to DNA. Science 263, 777–784 (1994).
Fuxreiter, M., Simon, I., Friedrich, P. & Tompa, P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026 (2004).
Sugase, K., Dyson, H. J. & Wright, P. E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007).
Shammas, S., Travis, A. J. & Clarke, J. Remarkably fast coupled folding and binding of the intrinsically disordered transactivation domain of cMyb to CBP KIX. J. Phys. Chem. B 117, 13346–13356 (2013).
Gianni, S., Morrone, A., Giri, R. & Brunori, M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochem. Biophys. Res. Commun. 428, 205–209 (2012).
Shammas, S. L., Travis, A. J. & Clarke, J. Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc. Natl Acad. Sci. USA 111, 12049–12054 (2014).
Rogers, J. M., Wong, C. T. & Clarke, J. Coupled folding and binding of the disordered protein PUMA does not require particular residual structure. J. Am. Chem. Soc. 136, 5197–5200 (2014).
Dogan, J., Schmidt, T., Mu, X., Engström, Å. & Jemth, P. Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. J. Biol. Chem. 287, 34316–34324 (2012).
Iešmantavicˇius, V., Dogan, J., Jemth, P., Teilum, K. & Kjaergaard, M. Helical propensity in an intrinsically disordered protein accelerates ligand binding. Angew. Chem. Int. Ed. Engl. (2014).
Schafmeister, C. E., Po, J. & Verdine, G. L. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc. 122, 5891–5892 (2000).
Bienkiewicz, E. A., Adkins, J. N. & Lumb, K. J. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27Kip1. Biochemistry 41, 752–759 (2002).
Baek, S. et al. Structure of the stapled p53 peptide bound to Mdm2. J. Am. Chem. Soc. 134, 103–106 (2011).
Otieno, S. & Kriwacki, R. Probing the role of nascent helicity in p27 function as a cell cycle regulator. PLoS ONE 7, e47177 (2012).
Borcherds, W. et al. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nature Chem. Biol. 10, 1000–1002 (2014).
Bertagna, A., Toptygin, D., Brand, L. & Barrick, D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem. Soc. Trans. 36, 157–166 (2008).
Wang, Y. et al. Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nature Chem. Biol. 7, 214–221 (2011).
Baker, J. M. R. et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nature Struct. Mol. Biol. 14, 738–745 (2007).
Mittag, T. et al. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure 18, 494–506 (2010).
Tompa, P. & Fuxreiter, M. Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 33, 2–8 (2008).
Ferreon, J. C., Martinez-Yamout, M. A., Dyson, H. J. & Wright, P. E. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc. Natl Acad. Sci. USA 106, 13260–13265 (2009).
Ishiyama, N. et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell–cell adhesion. Cell 141, 117–128 (2010).
Ferreon, A. C., Ferreon, J. C., Wright, P. E. & Deniz, A. A. Modulation of allostery by protein intrinsic disorder. Nature 498, 390–394 (2013).
Flock, T., Weatheritt, R. J., Latysheva, N. S. & Babu, M. M. Controlling entropy to tune the functions of intrinsically disordered regions. Curr. Opin. Struct. Biol. 26, 62–72 (2014).
Mukherjee, S. P. et al. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF κ B-driven transcription. PLoS Biol. 11, e1001647 (2013).
Pelka, P., Ablack, J. N. G., Fonseca, G. J., Yousef, A. F. & Mymryk, J. S. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82, 7252–7263 (2008).
Berk, A. J. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24, 7673–7685 (2005).
Fax, P., Lipinski, K. S., Esche, H. & Brockmann, D. cAMP-independent activation of the adenovirus type 12 E2 promoter correlates with the recruitment of CREB-1/ATF-1, E1A12S, and CBP to the E2-CRE. J. Biol. Chem. 275, 8911–8920 (2000).
Green, M., Panesar, N. K. & Loewenstein, P. M. The transcription-repression domain of the adenovirus E1A oncoprotein targets p300 at the promoter. Oncogene 27, 4446–4455 (2008).
Frye, J. J. et al. Electron microscopy structure of human APC/CCDH1–EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature Struct. Mol. Biol. 20, 827–835 (2013).
Nussinov, R. & Tsai, C. J. Allostery in disease and in drug discovery. Cell 153, 293–305 (2013).
Motlagh, H. N., Wrabl, J. O., Li, J. & Hilser, V. J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
Hilser, V. J. & Thompson, E. B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl Acad. Sci. USA 104, 8311–8315 (2007).
Garcia-Pino, A. et al. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142, 101–111 (2010).
Li, J., Motlagh, H. N., Chakuroff, C., Thompson, E. B. & Hilser, V. J. Thermodynamic dissection of the intrinsically disordered N-terminal domain of human glucocorticoid receptor. J. Biol. Chem. 287, 26777–26787 (2012).
Krishnan, N. et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nature Chem. Biol. 10, 558–566 (2014).
Pejaver, V. et al. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 23, 1077–1093 (2014).
Iakoucheva, L. M. et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 (2004).
Chrivia, J. C. et al. Phosphorylated CREB binds specifically to nuclear protein CBP. Nature 365, 855–859 (1993).
Radhakrishnan, I. et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91, 741–752 (1997).
Nash, P. et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001).
Mittag, T. et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl Acad. Sci. USA 105, 17772–17777 (2008).
Samaga, R. & Klamt, S. Modeling approaches for qualitative and semi-quantitative analysis of cellular signaling networks. Cell Commun. Signal. 11, 43 (2013).
Ferrell, J. E. Jr & Ha, S. H. Ultrasensitivity part II: multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem. Sci. 39, 556–569 (2014).
Lyons, N. A., Fonslow, B. R., Diedrich, J. K., Yates, J. R. & Morgan, D. O. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nature Struct. Mol. Biol. 20, 194–201 (2013).
Meek, D. W. & Anderson, C. W. Posttranslational Modification of p53: Cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950 (2009).
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Vousden, K. H. & Prives, C. Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009).
Grossman, S. R. et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300, 342 (2003).
Ferreon, J. C. et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc. Natl Acad. Sci. USA 106, 6591–6596 (2009).
Sakaguchi, K. et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275, 9278–9283 (2000).
Schon, O., Friedler, A., Freund, S. & Fersht, A. R. Binding of p53-derived ligands to MDM2 induces a variety of long range conformational changes. J. Mol. Biol. 336, 197–202 (2004).
Müller-Späth, S. et al. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl Acad. Sci. USA 107, 14609–14614 (2010).
Querfurth, C. et al. Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol. Cell 43, 713–722 (2011).
Baker, C. L., Kettenbach, A. N., Loros, J. J., Gerber, S. A. & Dunlap, J. C. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell 34, 354–363 (2009).
Tang, C. T. et al. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc. Natl Acad. Sci. USA 106, 10722–10727 (2009).
Holt, L. J. et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 (2009).
Kõivomägi, M. et al. Multisite phosphorylation networks as signal processors for Cdk1. Nature Struct. Mol. Biol. 20, 1415–1424 (2013).
Pufall, M. A. & Graves, B. J. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18, 421–462 (2002).
Trudeau, T. et al. Structure and intrinsic disorder in protein autoinhibition. Structure 21, 332–341 (2013).
Li, P., Martins, I. R. S., Amarasinghe, G. K. & Rosen, M. K. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nature Struct. Mol. Biol. 15, 613–618 (2008).
Yu, B. et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246–256 (2010).
Wu, H. Higher-order assemblies in a new paradigm of signal transduction. Cell 153, 287–292 (2013).
Weatheritt, R. J., Gibson, T. J. & Babu, M. M. Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nature Struct. Mol. Biol. 21, 833–839 (2014).
Li, J. et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 (2012).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Kedersha, N., Ivanov, P. & Anderson, P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 (2013).
Wippich, F. et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013).
Michelitsch, M. D. & Weissman, J. S. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc. Natl Acad. Sci. USA 97, 11910–11915 (2000).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Buljan, M. et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883 (2012).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein–protein interaction networks. Mol. Cell 46, 884–892 (2012).
Korneta, I. & Bujnicki, J. M. Intrinsic disorder in the human spliceosomal proteome. PLoS Computat. Biol. 8, e1002641 (2012).
Romero, P. R. et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA 103, 8390–8395 (2006).
Mermoud, J. E., Cohen, P. & Lamond, A. I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 20, 5263–5269 (1992).
Xiao, S. H. & Manley, J. L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344 (1997).
Xiang, S. et al. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure 21, 2162–2174 (2013).
Kwon, I. et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014).
Muñoz, M. J., de la Mata, M. & Kornblihtt, A. R. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 35, 497–504 (2010).
Hsin, J.-P. & Manley, J. L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012).
David, C. J., Boyne, A. R., Millhouse, S. R. & Manley, J. L. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65–Prp19 complex. Genes Dev. 25, 972–983 (2011).
Dye, M. J., Gromak, N. & Proudfoot, N. J. Exon tethering in transcription by RNA polymerase II. Mol. Cell 21, 849–859 (2006).
Theillet, F. X. et al. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem. Rev. 114, 6661–6714 (2014).
Sakon, J. J. & Weninger, K. R. Detecting the conformation of individual proteins in live cells. Nature Methods 7, 203–205 (2010).
Gebhardt, J. C. et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nature Methods 10, 421–426 (2013).
Wells, M. et al. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl Acad. Sci. USA 105, 5762–5767 (2008).
Cheng, Y. et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 24, 435–442 (2006).
Mészáros, B., Tompa, P., Simon, I. & Dosztányi, Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 372, 549–561 (2007).
Lao, B. B. et al. Rational design of topographical helix mimics as potent inhibitors of protein–protein interactions. J. Am. Chem. Soc. 136, 7877–7888 (2014).
Lao, B. B. et al. In vivo modulation of hypoxia-inducible signaling by topographical helix mimetics. Proc. Natl Acad. Sci. USA 111, 7588–7593 (2014).
Hammoudeh, D. I., Follis, A. V., Prochownik, E. V. & Metallo, S. J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-myc. J. Am. Chem. Soc. 131, 7390–7401 (2009).
Davey, N. E., Trave, G. & Gibson, T. J. How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169 (2011).
Hagai, T., Azia, A., Babu, M. M. & Andino, R. Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 7, 1729–1739 (2014).
Peng, K. et al. Optimizing intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 3, 35–60 (2005).
Dosztányi, Z., Csizmok, V., Tompa, P. & Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 (2005).
Dosztányi, Z., Csizmok, V., Tompa, P. & Simon, I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 (2005).
Dosztányi, Z., Mészáros, B. & Simon, I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25, 2745–2746 (2009).
Mészáros, B., Simon, I. & Dosztányi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 5, e1000376 (2009).
Obradovic, Z. et al. Predicting intrinsic disorder from amino acid sequence. Proteins: Struct. Function Bioinformat. 53 (Suppl. 6), 566–572 (2003).
Ishida, T. & Kinoshita, K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464 (2007).
Walsh, I., Martin, A. J., Di Domenico, T. & Tosatto, S. C. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics 28, 503–509 (2012).
Fukuchi, S., Homma, K., Minezaki, Y., Gojobori, T. & Nishikawa, K. Development of an accurate classification system of proteins into structured and unstructured regions that uncovers novel structural domains: its application to human transcription factors. BMC Struct. Biol. 9, 26 (2009).
Wojciak, J. M., Martinez-Yamout, M. A., Dyson, H. J. & Wright, P. E. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 28, 948–958 (2009).
Bösch, C., Bundi, A., Oppliger, M. & Wüthrich, K. 1H nuclear-magnetic-resonance studies of the molecular conformation of monomeric glucagon in aqueous solution. Eur. J. Biochem. 91, 209–214 (1978).
Altarejos, J. Y. & Montminy, M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature Rev. Mol. Cell Biol. 12, 141–151 (2011).
Sickmeier, M. et al. DisProt: the database of disordered proteins. Nucleic Acids Res. 35, D786–D793 (2007).
Acknowledgements
The authors apologize to the many colleagues whose work could not be cited owing to space limitations. Work in the authors' laboratories was supported by grants CA096865 (P.E.W.) and GM71862 (H.J.D.) from the US National Institutes of Health and by the Skaggs Institute for Chemical Biology (P.E.W.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Association rates
-
The speeds with which complexes are created from their component parts. The speed of the reverse process, detachment of the components, is known as the dissociation rate. The binding affinity is determined by the relative magnitude of the association and dissociation rates; for example, a fast association rate ('on-rate') and a slow dissociation rate ('off-rate') is characteristic of a high-affinity complex.
- Conformational ensembles
-
Structural descriptions of proteins that do not have a single, well-ordered three-dimensional structure. Conformational ensembles contain a multitude of different interconverting structures, which, when averaged, are consistent with observed parameters, such as nuclear magnetic resonance (NMR) spectra or small-angle X-ray scattering data.
- Binding free energy
-
The difference in free energy (ΔG) between the free and bound states of a complex. If the complex is stable, the binding free energy is negative.
Rights and permissions
About this article
Cite this article
Wright, P., Dyson, H. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16, 18–29 (2015). https://doi.org/10.1038/nrm3920
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3920
This article is cited by
-
Pathogenic mutations of human phosphorylation sites affect protein–protein interactions
Nature Communications (2024)
-
Structural biases in disordered proteins are prevalent in the cell
Nature Structural & Molecular Biology (2024)
-
Protein degradation by human 20S proteasomes elucidates the interplay between peptide hydrolysis and splicing
Nature Communications (2024)
-
Liquid–liquid phase separation in Alzheimer’s disease
Journal of Molecular Medicine (2024)
-
Relation between flexibility and intrinsically disorder regions in thermosensitive TRP channels reveal allosteric effects
European Biophysics Journal (2024)