Key Points
-
The human gut is home to dense bacterial and phage populations that are involved in regulating human health.
-
Phages regulate bacterial abundance, diversity and metabolism in numerous ecosystems, but their effects in the human gut remain largely unexplored.
-
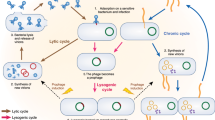
Despite high bacterial abundance and metabolism, the majority of described phages in the gut are integrated within their bacterial hosts, which suggests dynamic interactions that are specific to this system.
-
Different bacteria–phage interactions occur depending on the health status and development stage of the human host. Characterization of these interactions would provide unique ways to improve disease or developmental outcomes.
-
Further research on phage replication cycles and phage pharmacodynamics is essential before considering their therapeutic use for human health.
Abstract
The human gut is host to one of the densest microbial communities known, the gut microbiota, which contains bacteria, archaea, viruses, fungi and other microbial eukaryotes. Bacteriophages in the gut are largely unexplored, despite their potential to regulate bacterial communities and thus human health. In addition to helping us understand gut homeostasis, applying an ecological perspective to the study of bacterial and phage communities in the gut will help us to understand how this microbial system functions. For example, temporal studies of bacteria, phages and host immune cells in the gut during health and disease could provide key information about disease development and inform therapeutic treatments, whereas understanding the regulation of the replication cycles of phages could help harness the gut microbiota to improve disease outcomes. As the most abundant biological entities in our gut, we must consider bacteriophages in our pursuit of personalized medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rohwer, F., Prangishvili, D. & Lindell, D. Roles of viruses in the environment. Environ. Microbiol. 11, 2771–2774 (2009).
Mills, S. et al. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 4, 4–16 (2013).
De Smet, J. et al. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 10, 1823–1835 (2016).
Rodriguez-Valera, F. et al. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836 (2009).
Enault, F. et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247 (2017).
Parfrey, L. W., Walters, W. A. & Knight, R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2, 153 (2011).
Hoffmann, C. et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 8, e66019 (2013).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Hewitson, J. P. & Maizels, R. M. Vaccination against helminth parasite infections. Expert Rev. Vaccines 13, 473–487 (2014).
Tlaskalova-Hogenova, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8, 110–120 (2011).
Wilson, M. in Bacteriology of Humans: An Ecological Perspective 278–279 (Wiley-Blackwell, 2008).
Blaut, M. Ecology and physiology of the intestinal tract. Curr. Top. Microbiol. Immunol. 358, 247–272 (2013).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Sommer, F. & Backhed, F. The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006). A review that identifies possible ecological rules governing the diversity of the bacterial communities in the human gut.
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014). This study provides a comprehensive time-series analysis of gut and oral bacterial communities in two healthy individuals over the course of 1 year.
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015). This study details a comparison between the gut viromes of patients with IBD, showing disease-specific changes in virome diversity that are not explained by the changes in bacterial communities.
Reyes, A. et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl Acad. Sci. USA 112, 11941–11946 (2015).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Reyes, A., Wu, M., McNulty, N. P., Rohwer, F. L. & Gordon, J. I. Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc. Natl Acad. Sci. USA 110, 20236–20241 (2013). This study shows that mice that are colonized by a specific bacterial community isolated from the human gut and are infected by phages have reproducible and non-simultaneous bacterial infection patterns.
Zhang, T. et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4, e3 (2006).
Minot, S. et al. Rapid evolution of the human gut virome. Proc. Natl Acad. Sci. USA 110, 12450–12455 (2013). A temporal study of the healthy human gut virome over 2.5 years, which shows long-term stability of diversity in the gut virome and the rapid evolution of some long-term members of the gut virome.
Manrique, P. et al. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113, 10400–10405 (2016).
Browne, H. P. et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 (2016).
Myhrvold, C., Kotula, J. W., Hicks, W. M., Conway, N. J. & Silver, P. A. A distributed cell division counter reveals growth dynamics in the gut microbiota. Nat. Commun. 6, 10039 (2015).
Breitbart, M. et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159, 367–373 (2008).
Sharon, I. et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 23, 111–120 (2013).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011).
Lim, E. S. et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Thingstad, T. F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328 (2000).
Weinbauer, M. G. & Rassoulzadegan, F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11 (2004).
Mahana, D. et al. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med. 8, 48 (2016).
Charbonneau, M. R. et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164, 859–871 (2016).
Harrison, E., Laine, A. L., Hietala, M. & Brockhurst, M. A. Rapidly fluctuating environments constrain coevolutionary arms races by impeding selective sweeps. Proc. Biol. Sci. 280, 20130937 (2013).
Gomez, P. & Buckling, A. Bacteria–phage antagonistic coevolution in soil. Science 332, 106–109 (2011). This study in soil microcosms illustrates that bacteria–phage coevolution in soil leads to fluctuating selection dynamics between bacteria and phages.
Hall, A. R., Scanlan, P. D., Morgan, A. D. & Buckling, A. Host–parasite coevolutionary arms races give way to fluctuating selection. Ecol. Lett. 14, 635–642 (2011).
van Houte, S., Buckling, A. & Westra, E. R. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 80, 745–763 (2016).
Lopez Pascua, L. et al. Higher resources decrease fluctuating selection during host–parasite coevolution. Ecol. Lett. 17, 1380–1388 (2014).
Williamson, K. E., Radosevich, M., Smith, D. W. & Wommack, K. E. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 9, 2563–2574 (2007).
Weinbauer, M. G., Brettar, I. & Hofle, M. G. Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic marine waters. Limnol. Oceanogr. 48, 1457–1465 (2003).
Weinbauer, M. G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 (2004).
Silveira, C. B. & Rohwer, F. L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2, 16010 (2016).
Lenski, R. E. Dynamics of interactions between bacteria and virulent bacteriophage. Adv. Microb. Ecol. 10, 1–44 (1988).
Knowles, B. et al. Lytic to temperate switching of viral communities. Nature 531, 466–470 (2016). This study proposes the piggyback-the-winner model in host-associated microbial communities.
Touchon, M., Bernheim, A. & Rocha, E. P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754 (2016).
Peterson, D. A., Frank, D. N., Pace, N. R. & Gordon, J. I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 (2008).
Perez-Brocal, V. et al. Study of the viral and microbial communities associated with Crohn's disease: a metagenomic approach. Clin. Transl Gastroenterol. 4, e36 (2013).
Lepage, P. et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57, 424–425 (2008).
Brockhurst, M. A., Morgan, A. D., Fenton, A. & Buckling, A. Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens–Φ2 model system. Infect. Genet. Evol. 7, 547–552 (2007).
Maranger, R. & Bird, D. F. Viral abundance in aquatic systems — a comparison between marine and fresh-waters. Mar. Ecol. Prog. Ser. 121, 217–226 (1995).
Williamson, K. E., Radosevich, M. & Wommack, K. E. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71, 3119–3125 (2005).
Hodyra-Stefaniak, K. et al. Mammalian host-versus-phage immune response determines phage fate in vivo. Sci. Rep. 5, 14802 (2015).
Majewska, J. et al. Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 7, 4783–4799 (2015).
Dabrowska, K. et al. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 88, 12551–12557 (2014).
Miernikiewicz, P. et al. T4 phage tail adhesin Gp12 counteracts LPS-induced inflammation in vivo. Front. Microbiol. 7, 1112 (2016).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013). This study shows that phage immunoglobulin-like proteins enable increased phage adherence to mucosal surfaces of metazoan hosts, including human intestinal cells, and provide a first line of defence against bacterial pathogens.
Mogensen, T. H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273 (2009).
Fischbach, M. A. & Sonnenburg, J. L. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10, 336–347 (2011).
Rossmann, F. S. et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 11, e1004653 (2015).
Erez, Z. et al. Communication between viruses guides lysis–lysogeny decisions. Nature 541, 488–493 (2017). This study characterizes three phage genes that are involved in a phage-specific peptide communication system to coordinate the lysis–lysogeny decision of phages from the SPbeta group.
Jonczyk, E., Klak, M., Miedzybrodzki, R. & Gorski, A. The influence of external factors on bacteriophages — review. Folia Microbiol. (Praha) 56, 191–200 (2011).
Verthe, K., Possemiers, S., Boon, N., Vaneechoutte, M. & Verstraete, W. Stability and activity of an Enterobacter aerogenes-specific bacteriophage under simulated gastro-intestinal conditions. Appl. Microbiol. Biotechnol. 65, 465–472 (2004).
Ma, Y. et al. Microencapsulation of bacteriophage felix O1 into chitosan-alginate microspheres for oral delivery. Appl. Environ. Microbiol. 74, 4799–4805 (2008).
Maura, D., Galtier, M., Le Bouguenec, C. & Debarbieux, L. Virulent bacteriophages can target O104:H4 enteroaggregative Escherichia coli in the mouse intestine. Antimicrob. Agents Chemother. 56, 6235–6242 (2012).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Kim, M. S. & Bae, J. W. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ. Microbiol. 18, 1498–1510 (2016).
United Nations Children's Fund, World Health Organization & The World Bank. UNICEF–WHO–World Bank joint child malnutrition estimates. World Health Organization http://www.who.int/nutgrowthdb/jme_unicef_who_wb.pdf (2012).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013). This in vitro study identifies the rapid effects of therapeutic compounds on the gene expression, physiology and community structure of healthy gut bacterial communities, including the higher transcription of prophage induction genes.
Pavlova, S. I. & Tao, L. Induction of vaginal Lactobacillus phages by the cigarette smoke chemical benzo[a]pyrene diol epoxide. Mutat. Res. 466, 57–62 (2000).
Willner, D. et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4547–4553 (2011).
Taguer, M. & Maurice, C. F. The complex interplay of diet, xenobiotics, and microbial metabolism in the gut: implications for clinical outcomes. Clin. Pharmacol. Ther. 99, 588–599 (2016).
Mylon, S. E. et al. Influence of salts and natural organic matter on the stability of bacteriophage MS2. Langmuir 26, 1035–1042 (2010).
Lenzi, L. J., Lucchesi, P. M., Medico, L., Burgan, J. & Kruger, A. Effect of the food additives sodium citrate and disodium phosphate on shiga toxin-producing Escherichia coli and production of stx-phages and Shiga toxin. Front. Microbiol. 7, 992 (2016).
Allen, H. K. et al. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2, e00260-11 (2011).
DeMarini, D. M. & Lawrence, B. K. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat. Res. 267, 1–17 (1992).
Wegrzyn, G. & Wegrzyn, A. Genetic switches during bacteriophage λ development. Prog. Nucleic Acid Res. Mol. Biol. 79, 1–48 (2005).
Modi, S. R., Lee, H. H., Spina, C. S. & Collins, J. J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013).
Los, J. M., Los, M., Wegrzyn, G. & Wegrzyn, A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb. Pathog. 47, 289–298 (2009).
Volkova, V. V., Lu, Z., Besser, T. & Grohn, Y. T. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl. Environ. Microbiol. 80, 4350–4362 (2014).
Sarker, S. A. et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137 (2016).
Nailor, M. D. & Sobel, J. D. Antibiotics for Gram-positive bacterial infections: vancomycin, teicoplanin, quinupristin/dalfopristin, oxazolidinones, daptomycin, dalbavancin, and telavancin. Infect. Dis. Clin. North Am. 23, 965–982 (2009).
Ross, A., Ward, S. & Hyman, P. More is better: selecting for broad host range bacteriophages. Front. Microbiol. 7, 1352 (2016).
Bruttin, A. & Brussow, H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob. Agents Chemother. 49, 2874–2878 (2005).
Galtier, M. et al. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environ. Microbiol. 18, 2237–2245 (2016).
Wright, A., Hawkins, C. H., Anggard, E. E. & Harper, D. R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357 (2009).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02116010 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02664740 (2016).
Lu, T. K. & Collins, J. J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl Acad. Sci. USA 106, 4629–4634 (2009).
Libis, V. K. et al. Silencing of antibiotic resistance in E. coli with engineered phage bearing small regulatory RNAs. ACS Synth. Biol. 3, 1003–1006 (2014).
Chan, B. K. et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6, 26717 (2016).
Alang, N. & Kelly, C. R. Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2, ofv004 (2015).
Sheth, R. U., Cabral, V., Chen, S. P. & Wang, H. H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200 (2016).
Cooper, C., Khan Mirzaei, M. & Nilsson, A. S. Adapting drug approval pathways for bacteriophage-based therapeutics. Front. Microbiol. 7, 1209 (2016).
Nale, J. Y. et al. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob. Agents Chemother. 60, 968–981 (2016).
Abedon, S. T., Kuhl, S. J., Blasdel, B. G. & Kutter, E. M. Phage treatment of human infections. Bacteriophage 1, 66–85 (2011). A review of historical and contemporary research on the use of phages to treat human infections.
Manichanh, C., Borruel, N., Casellas, F. & Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608 (2012).
Pirnay, J. P. et al. The phage therapy paradigm: pret-a-porter or sur-mesure? Pharm. Res. 28, 934–937 (2011).
Nilsson, A. S. Phage therapy — constraints and possibilities. Ups. J. Med. Sci. 119, 192–198 (2014).
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389 (2005).
Bull, J. J. & Gill, J. J. The habits of highly effective phages: population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 5, 618 (2014).
Khan Mirzaei, M. & Nilsson, A. S. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10, e0118557 (2015).
Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Paul, J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2, 579–589 (2008). This study identifies phage-encoded repressors and transcriptional regulators of bacterial metabolism that enable the survival of the bacterial host in unfavourable environmental conditions in marine systems.
Brussow, H., Canchaya, C. & Hardt, W. D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602 (2004).
Smeal, S. W., Schmitt, M. A., Pereira, R. R., Prasad, A. & Fisk, J. D. Simulation of the M13 life cycle I: assembly of a genetically-structured deterministic chemical kinetic simulation. Virology 500, 259–274 (2017).
Cenens, W., Makumi, A., Mebrhatu, M. T., Lavigne, R. & Aertsen, A. Phage–host interactions during pseudolysogeny: lessons from the Pid/dgo interaction. Bacteriophage 3, e25029 (2013).
Feiner, R. et al. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650 (2015).
Reyesa, A. et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl Acad. Sci. USA 112, 11941–11946 (2015).
Thingstad, T. F. & Lignell, R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13, 19–27 (1997).
Maurice, C. F. et al. Disentangling the relative influence of bacterioplankton phylogeny and metabolism on lysogeny in reservoirs and lagoons. ISME J. 5, 831–842 (2011).
Bibby, K. Improved bacteriophage genome data is necessary for integrating viral and bacterial ecology. Microb. Ecol. 67, 242–244 (2014).
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Samson, J. E., Magadan, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687 (2013).
Stern, A. & Sorek, R. The phage–host arms race: shaping the evolution of microbes. Bioessays 33, 43–51 (2011).
Weitz, J. S., Hartman, H. & Levin, S. A. Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl Acad. Sci. USA 102, 9535–9540 (2005).
Acknowledgements
This work was supported by the Canada Research Chair Program and the Bill and Melinda Gates Foundation (OPP1139814). The authors thank members of the Maurice laboratory and E. Haggård-ljungquist for constructive comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Microbiome
-
The collective genomes of all microorganisms in an ecosystem.
- Microbiota
-
The collection of all microorganisms that exist in an ecosystem.
- Xenobiotics
-
Chemical substances that are foreign to an ecosystem.
- Dysbiosis
-
An imbalance in microbial diversity that is usually characterized by a decrease in specific members of the Firmicutes and an increase in members of the Proteobacteria.
- Genome annotation
-
The process of identifying, characterizing and annotating all coding and non-coding regions in a genome.
- Cryptic phages
-
Defective prophages that lack the ability to return to the lytic cycle.
- Kill-the-winner
-
(KTW). A specific case of Lotka–Volterra dynamics that is applied to bacterial and phage communities. The KTW hypothesis models the frequency-dependent viral infection of temporally abundant and active bacterial taxa, allowing for high bacterial diversity.
- Restriction–modification
-
(R–M). A bacterial defence system against invading foreign unmethylated DNA; for example, from phages and plasmids. Unmethylated DNA sites are recognized and cleaved by specific bacterial restriction endonucleases.
- Selective sweeps
-
The reduction or elimination of genetic variation in regions linked to a recently fixed beneficial mutation that increases host fitness.
- Fluctuating selection dynamics
-
A model of co-evolutionary dynamics that is characterized by the fluctuation of the selection pressure on genes, phenotypes or species in variable environments over time. In this model, there is maintenance of within-population genetic diversity over time, with variants that might become advantageous under appropriate environmental conditions.
- Piggyback-the-winner model
-
A recent model of bacteria–phage interactions, whereby phages integrate as prophages and undergo the lysogenic replication cycle instead of the lytic cycle when bacterial density and activity increase.
- Abortive infection system
-
(Abi system). Phage infection that leads to bacterial death and loss of the phage genome, without the production of any phages.
- Competitive exclusion
-
A mechanism whereby two (or more) related species that are competing for the same resources cannot stably coexist in the same environment and must specialize.
- Quorum sensing
-
A system of bacteria-produced molecular stimuli and responses that coordinate bacterial gene expression (of genes that are involved in biofilm formation, virulence and antibiotic resistance), according to the local density of the bacterial population.
- Gnotobiotic mice
-
Germ-free mice or antibiotic-treated mice that are colonized with predefined bacterial species or communities.
- Chronic otitis
-
A term that is used to describe various symptoms that result from the long-term damage of the middle ear by infection and inflammation.
- Pharmacodynamics
-
The study of the effects and mechanisms of action of a compound (typically therapeutic drugs) on a living organism.
- Pharmacokinetics
-
The study of the absorption, distribution, metabolism and excretion of a compound, typically applied to the study of therapeutic drugs.
- Temperate phages
-
Phages that replicate through lysogenic or lytic replication.
- Latency period
-
The time between phage infection and bacterial lysis with the release of new progeny.
- Burst size
-
The number of progeny produced per infecting phage.
Rights and permissions
About this article
Cite this article
Mirzaei, M., Maurice, C. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15, 397–408 (2017). https://doi.org/10.1038/nrmicro.2017.30
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.30
This article is cited by
-
Eco-evolutionary dynamics of gut phageome in wild gibbons (Hoolock tianxing) with seasonal diet variations
Nature Communications (2024)
-
Positive and negative aspects of bacteriophages and their immense role in the food chain
npj Science of Food (2024)
-
Microbial circadian clocks: host-microbe interplay in diel cycles
BMC Microbiology (2023)
-
Benzo[a]pyrene stress impacts adaptive strategies and ecological functions of earthworm intestinal viromes
The ISME Journal (2023)
-
Slow growing bacteria survive bacteriophage in isolation
ISME Communications (2023)