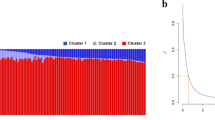
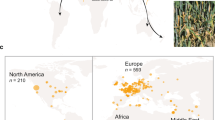
Key Points
-
During crop production, fungal plant pathogens cause severe yield losses. Uniform agricultural ecosystems are conducive for the rapid evolution and dissemination of pathogens.
-
The genomes of fungal plant pathogens can vary in size and composition, even between closely related species. Differences in the content of transposable elements cause variation in genome architecture.
-
Variation in genome architecture results from differences in population genetic factors, including effective population size and the strength of genetic drift.
-
During periods of low effective population size, non-adaptive mutations, such as transposable elements, can invade genomes and shape their architecture.
-
Transposable elements contribute to the establishment and maintenance of rapidly evolving genome compartments that can comprise virulence genes. High mutation rates in these compartments support the evolution of new virulence phenotypes.
Abstract
The fungal kingdom comprises some of the most devastating plant pathogens. Sequencing the genomes of fungal pathogens has shown a remarkable variability in genome size and architecture. Population genomic data enable us to understand the mechanisms and the history of changes in genome size and adaptive evolution in plant pathogens. Although transposable elements predominantly have negative effects on their host, fungal pathogens provide prominent examples of advantageous associations between rapidly evolving transposable elements and virulence genes that cause variation in virulence phenotypes. By providing homogeneous environments at large regional scales, managed ecosystems, such as modern agriculture, can be conducive for the rapid evolution and dispersal of pathogens. In this Review, we summarize key examples from fungal plant pathogen genomics and discuss evolutionary processes in pathogenic fungi in the context of molecular evolution, population genomics and agriculture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
31 August 2017
In Table 1 of this article, the species-specific hosts for both Leptosphaeria species should be crucifers (currently it is written as conifers). The mistakes have been corrected in the PDF and online. The authors apologize to the readers for any confusion caused.
References
Fisher, M., Henk, D. & Briggs, C. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194 (2012).
James, T. Y. et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822 (2006).
Lo Presti, L. et al. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 (2015).
Wolpert, T. J., Dunkle, L. D. & Ciuffetti, L. M. Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285 (2002).
Friesen, T. L., Faris, J. D., Solomon, P. S. & Oliver, R. P. Host-specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428 (2008).
Javelle, M., Vernoud, V., Rogowsky, P. M. & Ingram, G. C. Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39 (2011).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013).
Manning, V. A. & Ciuffetti, L. M. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 17, 3203–3212 (2005).
Djamei, A. et al. Metabolic priming by a secreted fungal effector. Nature 478, 395–398 (2011).
Bergelson, J., Kreitman, M., Stahl, E. A. & Tian, D. Evolutionary dynamics of plant R-genes. Science 292, 2281–2285 (2001).
Barrett, L. G. et al. Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 26, 2499–2513 (2009).
Dodds, P. N. et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA 103, 8888–8893 (2006).
Kanzaki, H. et al. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907 (2012).
Stukenbrock, E. H. et al. The making of a new pathogen: Insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 21, 2157–2166 (2011).
de Jonge, R. et al. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282 (2013).
Menardo, F. et al. Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat. Genet. 48, 201–205 (2016). This study provides an example of rapid adaptation to new host plants. The hybridization of two Blumeria graminis formae speciales resulted in the emergence of a new pathogen that infects a hybrid crop of wheat and rye.
Selmecki, A., Forche, A. & Berman, J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9, 991–1008 (2010).
Farrer, R. A. et al. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc. Natl Acad. Sci. USA 108, 18732–18736 (2011).
Raffaele, S. & Kamoun, S. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10, 417–430 (2012).
Seidl, M. F. & Thomma, B. P. Sex or no sex: evolutionary adaptation occurs regardless. Bioessays 36, 335–345 (2014).
Galazka, J. M. & Freitag, M. Variability of chromosome structure in pathogenic fungi — of 'ends and odds'. Curr. Opin. Microbiol. 20, 19–26 (2014).
Allen, R. L. et al. Host–parasite coevolutionary conflict between Arabidopsis and Downy Mildew. Science 306, 1957–1960 (2004).
Cao, J. et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43, 956–963 (2011).
Thrall, P. H. et al. Rapid genetic change underpins antagonistic coevolution in a natural host–pathogen metapopulation. Ecol. Lett. 15, 425–435 (2012).
Laine, A.-L., Burdon, J. J., Nemri, A. & Thrall, P. H. Host ecotype generates evolutionary and epidemiological divergence across a pathogen metapopulation. Proc. R. Soc. B Biol. Sci. 281, 20140522 (2014).
Persoons, A. et al. The escalatory Red Queen: population extinction and replacement following arms-race dynamics in poplar rust. Mol. Ecol. 26, 1902–1918 (2016).
Daverdin, G. et al. Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathog. 8, e1003020 (2012).
Rouxel, T. et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2, 202 (2011).
Brown, J. K. & Hovmøller, M. S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541 (2002).
Islam, M. T. et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14, 84 (2016).
Castroagudin, V. L. et al. Wheat blast disease caused by Pyricularia graminis-tritici sp. nov. Preprint at bioRxiv http://dx.doi.org/10.1101/051151 (2016).
Brasier, C. M. Rapid evolution of introduced plant pathogens via interspecific hybridization. Bioscience 51, 123 (2001).
Brasier, C. M. & Kirk, S. A. Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol. 59, 186–199 (2010).
McDonald, B. A. & Stukenbrock, E. H. Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Phil. Trans. R. Soc. B 371, 20160026 (2016).
Stukenbrock, E. H. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol. 199, 895–907 (2013).
Brown, J. K. & Tellier, A. Plant–parasite coevolution: bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 49, 345–367 (2011).
Hörger, A. C. et al. Balancing selection at the tomato RCR3 guardee gene family maintains variation in strength of pathogen defense. PLoS Genet. 8, e1002813 (2012).
Tellier, A., Moreno-Gámez, S. & Stephan, W. Speed of adaptation and genomic footprints of host–parasite coevolution under arms race and trench warfare dynamics. Evolution. 68, 2211–2224 (2014).
Van der Hoorn, R. A., De Wit, P. J. & Joosten, M. H. Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7, 67–71 (2002).
Badouin, H. et al. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Mol. Ecol. 26, 2041–2062 (2016).
Chuma, I. et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7, e1002147 (2011).
Ma, L.-J. et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373 (2010).
Faino, L. et al. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 26, 1091–1100 (2016). This study describes the importance of transposable elements in the formation of lineage-specific regions and their differential regulation in distinct genome compartments in V. dahliae.
Goodwin, S. B. et al. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070 (2011).
Coleman, J. J. et al. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5, e1000618 (2009).
Williams, A. H. et al. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17, 191 (2016).
Dutheil, J. Y. et al. A tale of genome compartmentalization: the evolution of virulence clusters in smut fungi. Genome Biol. Evol. 8, 681–704 (2016). This study provides details of the evolution of virulence-associated gene clusters in S. scitamineum that are driven by tandem gene duplication and transposable elements.
Dong, S., Raffaele, S. & Kamoun, S. The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65 (2015).
Stukenbrock, E. H. et al. Whole-genome and chromosome evolution associated with host adaptation and speciation of the wheat pathogen Mycosphaerella graminicola. PLoS Genet. 6, e1001189 (2010).
Houben, A., Banaei-Moghaddam, A. M., Klemme, S. & Timmis, J. N. Evolution and biology of supernumerary B chromosomes. Cell. Mol. Life Sci. 71, 467–478 (2014).
Miao, V. P., Covert, S. F. & VanEtten, H. D. A fungal gene for antibiotic resistance on a dispensable ('B') chromosome. Science 254, 1773 (1991).
Temporini, E. D. & VanEtten, H. D. Distribution of the pea pathogenicity (PEP) genes in the fungus Nectria haematococca mating population VI. Curr. Genet. 41, 107–114 (2002).
van der Does, H. C. et al. Transcription factors encoded on core and accessory chromosomes of Fusarium oxysporum induce expression of effector genes. PLoS Genet. 12, e1006401 (2016).
Wittenberg, A. H. J. et al. Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS ONE 4, e5863 (2009).
Croll, D., Zala, M. & McDonald, B. A. Breakage–fusion–bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 9, e1003567 (2013).
Stewart, E. l. et al. Quantitative trait locus mapping reveals complex genetic architecture of quantitative virulence in the wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. http://dx.doi.org/10.1111/mpp.12515 (2017).
Plissonneau, C. & Stürchler, A. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 7, e01231-16 (2016). In this study, comparison of the genome structure of two Z. tritici isolates identifies large chromosomal inversions and losses and/or gains of transposable element clusters, which highlights intraspecies genome diversity.
Chiara, M. et al. Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi. Genome Biol. Evol. 7, 3062–3069 (2015).
Soyer, J. L. et al. Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans. PLoS Genet. 10, e1004227 (2014). Effectors in L. maculans are located in AT-rich genome compartments. In this study, their regulation is shown to be mediated by the presence and absence of heterochromatin during infection.
Schotanus, K. et al. Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin 8, 1 (2015). In this study, Genome-wide comparison of histone methylation marks reveals distinct patterns of the facultative heterochromatin mark H3K27me3 on core and accessory chromosomes.
Kellner, R. et al. Expression profiling of the wheat pathogen Zymoseptoria tritici reveals genomic patterns of transcription and host-specific regulatory programs. Genome Biol. Evol. 6, 1353–1365 (2014).
Miao, V. P., Freitag, M. & Selker, E. U. Short TpA-rich segments of the ζ-η region induce DNA methylation in Neurospora crassa. J. Mol. Biol. 300, 249–273 (2000).
Tamaru, H. & Selker, E. U. Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol. Cell. Biol. 23, 2379–2394 (2003).
Fontanillas, E. et al. Degeneration of the non-recombining regions in the mating-type chromosomes of the anther-smut fungi. Mol. Biol. Evol. 32, 928–943 (2014).
Kämper, J. et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444, 97–101 (2006).
Schirawski, J. et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330, 1546–1548 (2010).
Croll, D., Lendenmann, M. H., Stewart, E. & McDonald, B. A. The impact of recombination hotspots on genome evolution of a fungal plant pathogen. Genetics 201, 1213–1228 (2015).
Ohm, R. A. et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8, e1003037 (2012).
Hane, J. K. et al. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 12, 1 (2011).
Grandaubert, J. et al. Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans–Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 15, 891 (2014). This study shows that the expansion of transposable elements in one member of the L. maculans–L. biglobosa species complex correlates to the evolution of pathogenicity in this species.
Chang, T.-C. et al. Comparative genomics of the sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLoS Genet. 12, e1005904 (2016).
Duplessis, S. et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl Acad. Sci. USA 108, 9166–9171 (2011).
Niehaus, E.-M. et al. Comparative 'omics' of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol. Evol. 8, 3574 (2016).
Lynch, M. & Walsh, B. The origins of genome architecture (Sinauer Associates Sunderland, 2007).
McDonald, B. A. & Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379 (2002).
Marais, G. & Charlesworth, B. Genome evolution: recombination speeds up adaptive evolution. Curr. Biol. 13, R68–R70 (2003).
Feldbrügge, M., Kämper, J., Steinberg, G. & Kahmann, R. Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7, 666–672 (2004).
Stukenbrock, E. H. & Dutheil, J. Y. Comparison of fine-scale recombination maps in fungal plant pathogens reveals dynamic recombination landscapes and intragenic hotspots. Preprint at bioRxiv https://dx.doi.org/10.1101/158907 (2017).
Zheng, Y. et al. Development of microsatellite markers and construction of genetic map in rice blast pathogen Magnaporthe grisea. Fungal Genet. Biol. 45, 1340–1347 (2008).
Talas, F. & McDonald, B. A. Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genomics 16, 996 (2015). This study provides details of the genomic structure of Fusarium graminearum field populations, which reveals a high degree of sexual recombination and gene flow that enables rapid adaptation to changing environments.
Muller, H. J. Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932).
Taylor, J. W., Jacobson, D. J. & Fisher, M. C. The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 37, 197–246 (1999).
Milgroom, M. G., del Mar Jiménez-Gasco, M., García, C. O., Drott, M. T. & Jiménez-Díaz, R. M. Recombination between clonal lineages of the asexual fungus Verticillium dahliae detected by genotyping by sequencing. PLoS ONE 9, e106740 (2014).
Short, D. P. G., Gurung, S., Hu, X., Inderbitzin, P. & Subbarao, K. V. Maintenance of sex-related genes and the co-occurrence of both mating types in Verticillium dahliae. PLoS ONE 9, e112145 (2014).
Ni, M. et al. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 11, e1001653 (2013).
Couch, B. C. et al. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170, 613–630 (2005).
Jin, Y., Szarbo, L. J. & Carson, M. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology 100, 432–435 (2010).
Xhaard, C. et al. The genetic structure of the plant pathogenic fungus Melampsora larici-populina on its wild host is extensively impacted by host domestication. Mol. Ecol. 20, 2739–2755 (2011).
Ali, S. et al. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PLoS Pathog. 10, e1003903 (2014).
Saleh, D. et al. Sex at the origin: an Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol. Ecol. 21, 1330–1344 (2012).
Ali, S., Leconte, M., Walker, A. S., Enjalbert, J. & Vallavieille-Pope, C. Reduction in the sex ability of worldwide clonal populations of Puccinia striiformis f.sp tritici. Fungal Genet. Biol. 47, 828–838 (2010).
Smith, K. M. et al. Epigenetics of filamentous fungi. Rev. Cell Biol. Mol. Med. http://dx.doi.org/10.1002/3527600906.mcb.201100035 (2012).
Dhillon, B., Cavaletto, J. R., Wood, K. V. & Goodwin, S. B. Accidental amplification and inactivation of a methyltransferase gene eliminates cytosine methylation in Mycosphaerella grami nicola. Genetics 186, 67–77 (2010).
Levin, H. L. & Moran, J. V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627 (2011).
Daboussi, M.-J. & Capy, P. Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 57, 275–299 (2003).
Capy, P., Gasperi, G., Biémont, C. & Bazin, C. Stress and transposable elements: co-evolution or useful parasites? Heredity (Edinb.) 85, 101–106 (2000).
Dolgin, E. S. & Charlesworth, B. The fate of transposable elements in asexual populations. Genetics 174, 817–827 (2006).
Lynch, M. & Conery, J. S. The origins of genome complexity. Science 302, 1401–1404 (2003).
Halkett, F. et al. Genetic discontinuities and disequilibria in recently established populations of the plant pathogenic fungus Mycosphaerella fijiensis. Mol. Ecol. 19, 3909–3923 (2010).
Munkacsi, A. B., Stoxen, S. & May, G. Ustilago maydis populations tracked maize through domestication and cultivation in the Americas. Proc. R. Soc. B Biol. Sci. 275, 1037–1046 (2008).
Whittle, C. A. & Johannesson, H. Evidence of the accumulation of allele-specific non-synonymous substitutions in the young region of recombination suppression within the mating-type chromosomes of Neurospora tetrasperma. Heredity (Edinb.) 107, 305–314 (2011).
Badouin, H. et al. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics 200, 1275–1284 (2015).
Perlin, M. H. et al. Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genomics 16, 1 (2015).
Okasha, S. Evolution and the Levels of Selection. (Oxford Univ. Press, 2006).
Anderson, J. B. & Kohn, L. M. in Sex in Fungi: Molecular Determination and Evolutionary Implications. (eds Heitman J. et al.)333–348 (ASM Press, 2007).
James, T. Y., Stenlid, J., Olson, Å. & Johannesson, H. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the Basidiomycete fungus Heterobasidion parviporum. Evolution. 62, 2279–2296 (2008).
Olson, A. & Stenlid, J. Plant pathogens: mitochondrial control of fungal hybrid virulence. Nature 411, 438 (2001).
Zheng, W. et al. High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 4, 2673 (2013).
Stukenbrock, E. H. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology 106, 104–112 (2016).
Stukenbrock, E. H., Christiansen, F. B., Hansen, T. T., Dutheil, J. Y. & Schierup, M. H. Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc. Natl Acad. Sci. USA 109, 10954–10959 (2012).
Inderbitzin, P., Davis, R. M., Bostock, R. M. & Subbarao, K. V. The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS ONE 6, e18260 (2011).
Shoji, J.-Y., Charlton, N. D., Yi, M., Young, C. A. & Craven, K. D. Vegetative hyphal fusion and subsequent nuclear behavior in Epichloë grass endophytes. PLoS ONE 10, e0121875 (2015).
Schardl, C. & Craven, K. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol. Ecol. 12, 2861–2873 (2003).
Baack, E. J. & Rieseberg, L. H. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518 (2007).
Richards, T. A. et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 21, 1897–1911 (2009).
Friesen, T. L. et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956 (2006).
Liu, Z. et al. The Tsn1–ToxA interaction in the wheat–Stagonospora nodorum pathosystem parallels that of the wheat–tan spot system. Genome 49, 1265–1273 (2006).
Faris, J. D. et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl Acad. Sci. USA 107, 13544–13549 (2010).
Mcdonald, M. C., Ahren, D., Simpfendorfer, S., Milgate, A. & Solomon, P. S. The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. http://dx.doi.org/10.1111/mpp.12535 (2017). This study shows that horizontal gene transfer of ToxA confers host-specific virulence across pathogens that infect wheat.
Nikolaidis, N., Doran, N. & Cosgrove, D. J. Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol. Biol. Evol. 31, 376–386 (2014).
de Jonge, R. et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl Acad. Sci. USA 109, 5110–5115 (2012).
Gardiner, D. M. et al. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8, e1002952 (2012).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Garsed, D. W. et al. The architecture and evolution of cancer neochromosomes. Cancer Cell 26, 653–667 (2014).
Wright, S. & Finnegan, D. Genome evolution: sex and the transposable element. Curr. Biol. 11, R296–R299 (2001).
Dang, Y., Yang, Q., Xue, Z. & Liu, Y. RNA interference in fungi: pathways, functions, and applications. Eukaryot. Cell 10, 1148–1155 (2011).
Cambareri, E. B., Jensen, B. C., Schabtach, E. & Selker, E. U. Repeat-induced G-C to A-T mutations in Neurospora. Science 244, 1571–1575 (1989).
Gladyshev, E. & Kleckner, N. Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat. Commun. 5, 3509 (2014).
Laurie, J. D. et al. Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24, 1733–1745 (2012). This study details a genome comparison of three closely related smut fungi that reveals transposable element expansion in Ustilago hordei and the loss of RNAi components in U. maydis.
Hood, M. E., Katawczik, M. & Giraud, T. Repeat-induced point mutation and the population structure of transposable elements in Microbotryum violaceum. Genetics 170, 1081–1089 (2005).
Idnurm, A. & Howlett, B. J. Analysis of loss of pathogenicity mutants reveals that repeat-induced point mutations can occur in the Dothideomycete Leptosphaeria maculans. Fungal Genet. Biol. 39, 31–37 (2003).
Zemach, A., McDaniel, I. E., Silva, P. & Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010).
Galagan, J. E. & Selker, E. U. RIP: The evolutionary cost of genome defense. Trends Genet. 20, 417–423 (2004).
Studt, L. et al. Knock-down of the methyltransferase Kmt6 relieves H3K27me3 and results in induction of cryptic and otherwise silent secondary metabolite gene clusters in Fusarium fujikuroi. Environ. Microbiol. 18, 4037–4054 (2016).
Connolly, L. R., Smith, K. M. & Freitag, M. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 9, e1003916 (2013).
Letunic, I. & Bork, P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007).
Grandaubert, J., Bhattacharyya, A. & Stukenbrock, E. H. RNA-seq based gene annotation and comparative genomics of four fungal grass pathogens in the genus Zymoseptoria identify novel orphan genes and species-specific invasions of transposable elements. G3 (Bethesda) 5, 1323–1333 (2015).
Vanheule, A. et al. Living apart together: crosstalk between the core and supernumerary genomes in a fungal plant pathogen. BMC Genomics 17, 670 (2016).
Spanu, P. D. et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330, 1543–1546 (2010).
Wicker, T. et al. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096 (2013).
Hacquard, S. et al. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl Acad. Sci. USA 110, E2219–E2228 (2013).
Dean, R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986 (2005).
Forgetta, V. et al. Sequencing of the Dutch elm disease fungus genome using the Roche/454 GS-FLX Titanium System in a comparison of multiple genomics core facilities. J. Biomol. Tech. 24, 39–49 (2013).
Cuomo, C. A. et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317, 1400–1402 (2007).
Acknowledgements
The authors thank B. McDonald, M. Freitag, J. Haueisen and J. Dutheil for helpful discussions and comments in regard to a previous version of this Review. Research carried out in the group of E.H.S. is funded by the Max Planck Society, Germany, and a personal grant from the State of Schleswig-Holstein, Germany, to E.H.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- Biotrophs
-
Pathogens that manipulate host defences and obtain their nutrients from living plant cells through specialized 'feeding' structures or hyphae formed intracellularly in the host.
- Necrotrophs
-
Pathogens that kill host cells by secreting toxins and enzymes to obtain nutrients for growth and reproduction.
- Hemibiotrophs
-
Pathogens that undergo a longer latent or biotrophic phase followed by a switch to necrotrophic growth.
- Effectors
-
Pathogen-produced molecules that are secreted during infection to manipulate host defences and facilitate pathogen invasion.
- Apoplastic space
-
Space outside of the plant plasma membrane.
- Symplastic space
-
Space on the inner side of the plant plasma membrane.
- Cultivars
-
Varieties of crop plants of the same species that have distinct phenotypes and genotypes. Distinct cultivars are obtained by plant breeding, whereby desired traits are selected and propagated.
- Transposable elements
-
Genetic elements that move from one location in the genome of their host to another. Transposable elements are also known as transposons.
- Metapopulation dynamics
-
Local fluctuations in the (actual as well as effective) population size of spatially separated populations that belong to the same species.
- Non-synonymous mutations
-
Nucleotide changes in coding sequences that alter the amino acid sequence in the translated proteins.
- Synonymous mutations
-
Nucleotide changes in coding sequences that alter the codons, but not the amino acid sequence, in translated proteins.
- Trench warfare
-
A model that predicts constant diversity in a host–pathogen system, due to the maintenance of multiple alleles at a co-evolving locus by positive diversifying selection or balancing selection.
- 'Arms race' evolution
-
The co-evolution of host and pathogen alleles, which results in the recurrent fixation of advantageous alleles at a co-evolving locus. The fixation of advantageous alleles is mediated by positive directional selection.
- Tajimas D
-
A statistical test parameter that is used in population genetics and DNA sequence analyses. Tajima's test is used to identify sequences that do not fit the neutral theory model, by which the fate of any mutation is determined by genetic drift.
- Divergence patterns
-
The distribution of substitutions varies along the genome, as different parts of the genome evolve by processes and by different rates. The underlying pattern of divergence can be investigated to unravel the history of mutational events and the effect of selection versus neutral processes on the sequence evolution.
- Selective sweep
-
An increase in the frequency of an advantageous allele (and closely linked chromosomal segments) that is caused by positive selection. Sweeps initially decrease genetic variation and subsequently lead to a local excess of rare alleles (homozygosity excess) as new unique mutations accumulate.
- Linkage disequilibrium
-
The non-random association of alleles at different loci.
- Accessory chromosomes
-
Chromosomes that are not present in all isolates of the same pathogenic species. Such chromosomes often encode determinants of host specificity.
- Heterothallic
-
In heterothallic species, two individuals that have opposite mating types are required for sexual reproduction. By contrast, homothallic organisms have both mating types in one thallus.
- Mesosynteny
-
A term that refers to the conservation of gene content on chromosomes, but a variation in the gene order and orientation. This is a phenomenon that is, thus far, particularly described in dothideomycete fungi.
- Effective population size
-
(Ne). The approximate number of breeding individuals that produce offspring that live to reproductive age. This number influences the rate of loss of genetic variation, the efficiency of natural selection, and the accumulation of beneficial and deleterious mutations. It is frequently much smaller than the number of individuals in a population.
- Sexual spores
-
Spores that originate from sexual crossings that differ morphologically from asexual spores.
- Muller's ratchet
-
The irreversible accumulation of deleterious mutations in organisms that reproduce asexually.
- Parasexual
-
A process whereby genetic material is exchanged between fused hyphae or cells without meiosis. Parasexuality enables the organism to recombine its genome and generate new genotypes in the absence of sexual mating.
- Selection coefficient
-
The average proportional reduction in fitness of one genotype relative to another owing to selection (designated by 's').
- Heterokaryons
-
Cells that contain two or more genetically distinct nuclei.
- Introgressive hybridization
-
The transfer of genes from one species to another through hybridization followed by backcrossing with the parental species.
Rights and permissions
About this article
Cite this article
Möller, M., Stukenbrock, E. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol 15, 756–771 (2017). https://doi.org/10.1038/nrmicro.2017.76
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.76
This article is cited by
-
Analysis of five near-complete genome assemblies of the tomato pathogen Cladosporium fulvum uncovers additional accessory chromosomes and structural variations induced by transposable elements effecting the loss of avirulence genes
BMC Biology (2024)
-
Quantitative pathogenicity and host adaptation in a fungal plant pathogen revealed by whole-genome sequencing
Nature Communications (2024)
-
Sequence and assembly of the genome of Seiridium unicorne, isolate CBS 538.82, causal agent of cypress canker disease
Journal of Plant Pathology (2024)
-
Enhanced oxidative stress resistance in Ustilago maydis and its implications on the virulence
International Microbiology (2024)
-
Current trends, limitations and future research in the fungi?
Fungal Diversity (2024)