Key Points
-
A common feature of host-adapted bacterial pathogens is the high-frequency, random on-and-off switching of surface proteins and glycans. This process, called phase variation, generates a diverse bacterial population and facilitates immune evasion and adaptation to host micro-environments. The most common mechanism of phase-variable gene expression is a tract of simple tandem repeats that mutate at high frequency, altering gene expression through frameshift mutations or changes in the promoter region.
-
Type III restriction–modification (R–M) systems are composed of two enzymes: a methyltransferase (encoded by a mod gene) and a restriction endonuclease (encoded by a res gene). Mod catalyses the methylation of a specific DNA recognition sequence, distinguishing 'self' DNA and protecting it from cleavage. Mod can act independently of the restriction enzyme and methylates one strand only. Res catalyses the double-stranded cleavage of unmethylated, 'non-self' DNA, but can only work as a complex with Mod, which contains the DNA recognition domain.
-
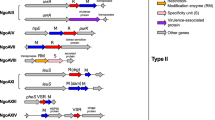
Numerous type III R–M systems in host-adapted bacterial pathogens contain repetitive motifs, indicating the potential for phase variation of these systems in many species, including Haemophilus influenzae, Helicobacter pylori, Mannheimia haemolytica, Neisseria meningitidis, Neisseria gonorrhoeae and Moraxella catarrhalis. Methyltransferases from type III R–M systems, unlike most other phase-variable genes, are not involved in the biosynthesis of surface antigens. R–M systems are traditionally involved in protection of the bacterial cell from incoming foreign DNA.
-
Several roles have been suggested for phase-variable type III R–M systems, including: acting as a barrier to bacteriophage or genetic exchange by transformation; allowing the release of DNA into the environment for uptake by other cells in the population, through autolytic self-DNA degradation by the cognate restriction enzyme; and influencing gene regulation through differential methylation of the genome. A phase-variable methyltransferase could play a part in pathogenicity by randomizing virulence factor expression through global changes in methylation.
-
In DNA adenine methylase (Dam)-dependent phase variation systems, competition between Dam and a DNA-binding protein forms a DNA methylation pattern that controls gene expression at a target site. The target site's methylation state affects the DNA binding of a regulatory protein, which directly regulates transcription. The Dam methylase itself does not phase vary.
-
The 'phasevarion' (phase-variable regulon) is a novel genetic system coordinating the random switching of multiple genes. The mod genes undergo phase variation through their tandem repeats, and this in turn influences the random switching of the expression of multiple genes. The wide distribution of phase-variable mod genes indicates that this may be a common strategy used by host-adapted bacterial pathogens for the generation of 'differentiated' cell types with distinct niche specializations.
Abstract
In several host-adapted pathogens, phase variation has been found to occur in genes that encode methyltransferases associated with type III restriction–modification systems. It was recently shown that in the human pathogens Haemophilus influenzae, Neisseria gonorrhoeae and Neisseria meningitidis phase variation of a type III DNA methyltransferase, encoded by members of the mod gene family, regulates the expression of multiple genes. This novel genetic system has been termed the 'phasevarion' (phase-variable regulon). The wide distribution of phase-variable mod family genes indicates that this may be a common strategy used by host-adapted bacterial pathogens to randomly switch between distinct cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
van Ham, S. M., van Alphen, L., Mooi, F. R. and van Putten, J. P. Phase variation of Haemophilus influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell 73, 1187–1196 (1993).
Weiser, J. N., Williams, A. & Moxon, E. R. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 58, 3455–3457 (1990).
Hallet, B. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr. Opin. Microbiol. 4, 570–581 (2001). A comprehensive review focusing on the different phase variation mechanisms in bacteria.
Jennings, M. P., Hood, D. W., Peak, I. R., Virji, M. & Moxon, E. R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol. Microbiol. 18, 729–740 (1995).
van der Ende, A. et al. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the -10 and -35 regions of the promoter. J. Bacteriol. 177, 2475–2480 (1995).
Moxon, E. R. & Thaler, D. S. Microbial genetics. The tinkerer's evolving tool-box. Nature 387, 659–662 (1997).
Robertson, B. D. & Meyer, T. F. Genetic variation in pathogenic bacteria. Trends Genet. 8, 422–427 (1992).
van der Woude, M. W. & Baumler, A. J. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17, 581–611 (2004). An excellent review of type I R–M systems.
van der Woude, M. W. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254, 190–197 (2006).
Moxon, R., Bayliss, C. & Hood, D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40, 307–333 (2006). A detailed review focusing on the role of simple tandem repeats in bacterial adaptation.
Levinson, G. & Gutman, G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203–221 (1987). An overview of the bacterial proteins and structures that are under the control of phase variation.
Chandler, M. & Fayet, O. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7, 497–503 (1993).
van Belkum, A., Scherer, S., van Alphen, L. & Verbrugh, H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62, 275–293 (1998).
Boyer, H. W. DNA restrictions and modification mechanisms in bacteria. Annu. Rev. Microbiol. 25, 153–176 (1971).
Bickle, T. A., Brack, C. & Yuan, R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc. Natl Acad. Sci. USA 75, 3099–3103 (1978).
Sears, A., Peakman, L. J., Wilson, G. G. & Szczelkun, M. D. Characterization of the Type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 33, 4775–4787 (2005).
Arber, W. & Dussoix, D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage λ. J. Mol. Biol. 5, 18–36 (1962).
Arber, W. & Wauters-Willems, D. Host specificity of DNA produced by Escherichia coli. XII. The two restriction and modification systems of strain 15T-. Mol. Gen. Genet. 108, 203–217 (1970).
Piekarowicz, A., Kauc, L. & Glover, S. W. Host specificity of DNA in Haemophilus influenzae: the restriction and modification systems in strains Rb and Rf. J. Gen. Microbiol. 81, 391–403 (1974).
Bullas, L. R., Colson, C. & Neufeld, B. Deoxyribonucleic acid restriction and modification systems in Salmonella: chromosomally located systems of different serotypes. J. Bacteriol. 141, 275–292 (1980).
Bourniquel, A. A. & Bickle, T. A. Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84, 1047–1059 (2002).
Dryden, D. T., Murray, N. E. & Rao, D. N. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 29, 3728–3741 (2001). A detailed description of the molecular mechanisms of DNA translocation and cleavage in type I and type III R–M systems.
Hadi, S. M., Bachi, B., Iida, S. & Bickle, T. A. DNA restriction–modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J. Mol. Biol. 165, 19–34 (1983).
Iida, S. et al. DNA restriction-modification genes of phage P1 and plasmid p15B. Structure and in vitro transciption. J. Mol. Biol. 165, 1–18 (1983).
Meisel, A., Bickle, T. A., Kruger, D. H. & Schroeder, C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature 355, 467–469 (1992).
Bachi, B., Reiser, J. & Pirrotta, V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J. Mol. Biol. 128, 143–163 (1979).
Janscak, P., Sandmeier, U., Szczelkun, M. D. & Bickle, T. A. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 306, 417–431 (2001).
Bheemanaik, S., Reddy, Y. V. & Rao, D. N. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J. 399, 177–190 (2006).
Sistla, S. & Rao, D. N. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 1–19 (2004). A classic review, covering type I, type II and type III R–M systems.
Roberts, R. J. & Cheng, X. Base flipping. Annu. Rev. Biochem. 67, 181–198 (1998).
Reddy, Y. V. & Rao, D. N. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 298, 597–610 (2000).
Liebert, K., Hermann, A., Schlickenrieder, M. & Jeltsch, A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 341, 443–454 (2004).
Ahmad, I. & Rao, D. N. Interaction of EcoP15I DNA methyltransferase with oligonucleotides containing the asymmetric sequence 5′-CAGCAG-3′. J. Mol. Biol. 242, 378–388 (1994).
Malone, T., Blumenthal, R. M. & Cheng, X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253, 618–632 (1995).
Humbelin, M. et al. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 200, 23–29 (1988).
Gorbalenya, A. E. & Koonin, E. V. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 291, 277–281 (1991).
Saha, S. & Rao, D. N. Mutations in the Res subunit of the EcoPI restriction enzyme that affect ATP-dependent reactions. J. Mol. Biol. 269, 342–354 (1997).
Saha, S., Ahmad, I., Reddy, Y. V., Krishnamurthy, V. & Rao, D. N. Functional analysis of conserved motifs in type III restriction-modification enzymes. Biol. Chem. 379, 511–517 (1998).
Pingoud, A. & Jeltsch, A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 246, 1–22 (1997).
Meselson, M. & Yuan, R. DNA restriction enzyme from E. coli. Nature 217, 1110–1114 (1968).
Haberman, A. The bacteriophage P1 restriction endonuclease. J. Mol. Biol. 89, 545–563 (1974).
Reiser, J. & Yuan, R. Purification and properties of the P15 specific restriction endonuclease from Escherichia coli. J. Biol. Chem. 252, 451–456 (1977).
Hadi, S. M. et al. DNA recognition and cleavage by the EcoP15 restriction endonuclease. J. Mol. Biol. 134, 655–666 (1979).
Ahmad, I., Krishnamurthy, V. & Rao, D. N. DNA recognition by the EcoP15I and EcoPI modification methyltransferases. Gene 157, 143–147 (1995).
Meisel, A., Mackeldanz, P., Bickle, T. A., Kruger, D. H. & Schroeder, C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 14, 2958–2966 (1995). A paper focusing on type III restriction endonucleases.
Bist, P. et al. S-adenosyl-L-methionine is required for DNA cleavage by type III restriction enzymes. J. Mol. Biol. 310, 93–109 (2001).
Peakman, L. J., Antognozzi, M., Bickle, T. A., Janscak, P. & Szczelkun, M. D. S-adenosyl methionine prevents promiscuous DNA cleavage by the EcoP1I type III restriction enzyme. J. Mol. Biol. 333, 321–335 (2003).
Szczelkun, M. D. How do proteins move along DNA? Lessons from type-I and type-III restriction endonucleases. Essays Biochem. 35, 131–143 (2000). A thorough explanation of the tracking mechanisms of the type I and type III R–M enzymes.
Murray, N. E., Daniel, A. S., Cowan, G. M. & Sharp, P. M. Conservation of motifs within the unusually variable polypeptide sequences of type I restriction and modification enzymes. Mol. Microbiol. 9, 133–143 (1993).
Mucke, M., Reich, S., Moncke-Buchner, E., Reuter, M. & Kruger, D. H. DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites. J. Mol. Biol. 312, 687–698 (2001).
Peakman, L. J. & Szczelkun, M. D. DNA communications by Type III restriction endonucleases—confirmation of 1D translocation over 3D looping. Nucleic Acids Res. 32, 4166–4174 (2004).
Reich, S., Gossl, I., Reuter, M., Rabe, J. P. & Kruger, D. H. Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I. J. Mol. Biol. 341, 337–343 (2004).
Crampton, N. et al. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl Acad. Sci. USA 104, 12755–12760 (2007).
Crampton, N. et al. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 26, 3815–3825 (2007).
Ramanathan, S. P. et al. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc. Natl Acad. Sci. USA 106, 1748–1753 (2009).
Tock, M. R. & Dryden, D. T. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472 (2005).
Zaleski, P., Wojciechowski, M. & Piekarowicz, A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology 151, 3361–3369 (2005).
Highlander, S. K. & Garza, O. The restriction-modification system of Pasteurella haemolytica is a member of a new family of type I enzymes. Gene 178, 89–96 (1996).
Dybvig, K., Sitaraman, R. & French, C. T. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl Acad. Sci. USA 95, 13923–13928 (1998).
Ryan, K. A. & Lo, R. Y. Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res. 27, 1505–1511 (1999).
De Bolle, X. et al. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35, 211–222 (2000). A paper detailing phase variation of a type III methyltransferase in H. influenzae.
de Vries, N. et al. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 184, 6615–6623 (2002).
Srikhanta, Y. N. et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 5, e1000400 (2009). Phasevarions in pathogenic Neisseria spp.
Saunders, N. J., Peden, J. F., Hood, D. W. & Moxon, E. R. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 27, 1091–1098 (1998).
Salaun, L., Linz, B., Suerbaum, S. & Saunders, N. J. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology 150, 817–830 (2004).
Fox, K. L., Srikhanta, Y. N. & Jennings, M. P. Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol. Microbiol. 65, 1375–1379 (2007).
Rocha, E. P. & Blanchard, A. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30, 2031–2042 (2002).
Hood, D. W. et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl Acad. Sci. USA 93, 11121–11125 (1996).
Seib, K. L., Peak, I. R. & Jennings, M. P. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 32, 159–165 (2002).
Fox, K. L. et al. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 35, 5242–5252 (2007).
Bayliss, C. D., Callaghan, M. J. & Moxon, E. R. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 34, 4046–4059 (2006).
Kroll, J. S., Wilks, K. E., Farrant, J. L. & Langford, P. R. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl Acad. Sci. USA 95, 12381–12385 (1998).
Ando, T. et al. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37, 1052–1065 (2000).
Donahue, J. P., Israel, D. A., Peek, R. M., Blaser, M. J. & Miller, G. G. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37, 1066–1074 (2000).
Gibbs, C. P. et al. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature 338, 651–652 (1989).
Hamilton, H. L. & Dillard, J. P. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59, 376–385 (2006).
Buhler, R. & Yuan, R. Characterization of a restriction enzyme from Escherichia coli K carrying a mutation in the modification subunit. J. Biol. Chem. 253, 6756–6760 (1978).
Willcock, D. F., Dryden, D. T. & Murray, N. E. A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J. 13, 3902–3908 (1994).
Doronina, V. A. & Murray, N. E. The proteolytic control of restriction activity in Escherichia coli K-12. Mol. Microbiol. 39, 416–428 (2001).
Makovets, S., Doronina, V. A. & Murray, N. E. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc. Natl Acad. Sci. USA 96, 9757–9762 (1999).
Murray, N. E. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64, 412–434 (2000).
Redaschi, N. & Bickle, T. A. Posttranscriptional regulation of EcoP1I and EcoP15I restriction activity. J. Mol. Biol. 257, 790–803 (1996).
Arber, W., Yuan, R. & Bickle, T. A. Strain-specific modification and restriction of DNA in bacteria. FEBS Proc. Symp. 9, 3–22 (1975).
Arber, W. DNA modification and restriction. Prog. Nucleic Acid Res. Mol. Biol. 14, 1–37 (1974).
De Backer, O. & Colson, C. Transfer of the genes for the StyLTI restriction-modification system of Salmonella typhimurium to strains lacking modification ability results in death of the recipient cells and degradation of their DNA. J. Bacteriol. 173, 1328–1330 (1991).
Sears, A. & Szczelkun, M. D. Subunit assembly modulates the activities of the Type III restriction-modification enzyme PstII in vitro. Nucleic Acids Res. 33, 4788–4796 (2005).
Blyn, L. B., Braaten, B. A. & Low, D. A. Regulation of pap pilin phase variation by a mechanism involving differential Dam methylation states. EMBO J. 9, 4045–4054 (1990).
Haagmans, W. & van der Woude, M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35, 877–887 (2000).
Nicholson, B. & Low, D. DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol. Microbiol. 35, 728–742 (2000).
Casadesus, J. & Low, D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 (2006).
Wion, D. & Casadesus, J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nature Rev. Microbiol. 4, 183–192 (2006). This and reference 90 present comprehensive reviews of epigenetic gene regulation in bacteria.
Marinus, M. G. & Casadesus, J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33, 488–503 (2009). An excellent review of the role of Dam methylation in host–pathogen interactions.
Heithoff, D. M., Sinsheimer, R. L., Low, D. A. & Mahan, M. J. An essential role for DNA adenine methylation in bacterial virulence. Science 284, 967–970 (1999). A detailed description of the essential role for DNA adenine methylation in bacterial virulence.
Garcia-Del Portillo, F., Pucciarelli, M. G. & Casadesus, J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl Acad. Sci. USA 96, 11578–11583 (1999).
Heithoff, D. M. et al. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 69, 6725–6730 (2001).
Pucciarelli, M. G., Prieto, A. I., Casadesus, J. & Garcia-del Portillo, F. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148, 1171–1182 (2002).
Balbontin, R. et al. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 8160–8168 (2006).
Prieto, A. I. et al. The GATC-binding protein SeqA is required for bile resistance and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 189, 8496–8502 (2007).
Jakomin, M., Chessa, D., Baumler, A. J. & Casadesus, J. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190, 7406–7413 (2008).
Chatti, A., Daghfous, D. & Landoulsi, A. Effect of repeated in vivo passage (in mice) on Salmonella typhimurium dam mutant virulence and fitness. Pathol. Biol. (Paris) 56, 121–124 (2008).
Chen, L. et al. Alteration of DNA adenine methylase (Dam) activity in Pasteurella multocida causes increased spontaneous mutation frequency and attenuation in mice. Microbiology 149, 2283–2290 (2003).
Julio, S. M. et al. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69, 7610–7615 (2001).
Julio, S. M., Heithoff, D. M., Sinsheimer, R. L., Low, D. A. & Mahan, M. J. DNA adenine methylase overproduction in Yersinia pseudotuberculosis alters YopE expression and secretion and host immune responses to infection. Infect. Immun. 70, 1006–1009 (2002).
Taylor, V. L., Titball, R. W. & Oyston, P. C. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology 151, 1919–1926 (2005).
Campellone, K. G. et al. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 63, 1468–1481 (2007).
Falker, S., Schilling, J., Schmidt, M. A. & Heusipp, G. Overproduction of DNA adenine methyltransferase alters motility, invasion, and the lipopolysaccharide O-antigen composition of Yersinia enterocolitica. Infect. Immun. 75, 4990–4997 (2007).
Mehling, J. S., Lavender, H. & Clegg, S. A Dam methylation mutant of Klebsiella pneumoniae is partially attenuated. FEMS Microbiol. Lett. 268, 187–193 (2007).
Kim, J. S. et al. Role of the Campylobacter jejuni Cj1461 DNA methyltransferase in regulating virulence characteristics. J. Bacteriol. 190, 6524–6529 (2008).
Murphy, K. C., Ritchie, J. M., Waldor, M. K., Lobner-Olesen, A. & Marinus, M. G. Dam methyltransferase is required for stable lysogeny of the Shiga toxin (Stx2)-encoding bacteriophage 933W of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 190, 438–441 (2008).
Low, D. A., Weyand, N. J. & Mahan, M. J. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69, 7197–7204 (2001).
Nou, X., Braaten, B., Kaltenbach, L. & Low, D. A. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14, 5785–5797 (1995).
Weyand, N. J. & Low, D. A. Regulation of Pap phase variation. Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J. Biol. Chem. 275, 3192–3200 (2000).
Henderson, I. R. & Owen, P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181, 2132–2141 (1999).
Danese, P. N., Pratt, L. A., Dove, S. L. & Kolter, R. The outer membrane protein, antigen 43 mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37, 424–432 (2000).
Srikhanta, Y. N., Maguire, T. L., Stacey, K. J., Grimmond, S. M. & Jennings, M. P. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl Acad. Sci. USA 102, 5547–5551 (2005). The first report of the phasevarion.
Hartmann, E., Lingwood, C. A. & Reidl, J. Heat-inducible surface stress protein (Hsp70) mediates sulphatide recognition of the respiratory pathogen Haemophilus influenzae. Infect. Immun. 69, 3438–3441 (2001).
Hartmann, E. & Lingwood, C. Brief heat shock treatment induces a long-lasting alteration in the glycolipid receptor binding specificity and growth rate of Haemophilus influenzae. Infect. Immun. 65, 1729–1733 (1997).
Herbert, M. A. et al. Signature Tagged Mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb. Pathog. 33, 211–223 (2002).
Pettersson, A., van der Ley, P., Poolman, J. T. & Tommassen, J. Molecular characterization of the 98-kilodalton iron-regulated outer membrane protein of Neisseria meningitidis. Infect. Immun. 61, 4724–4733 (1993).
Pettersson, A., Prinz, T., Umar, A., van der Biezen, J. & Tommassen, J. Molecular characterization of LbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol. Microbiol 27, 599–610 (1998).
Lin, L. F., Posfai, J., Roberts, R. J. & Kong, H. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA 98, 2740–2745 (2001).
Skoglund, A. et al. Functional analysis of the, M.HpyAIV DNA-methyltransferase of Helicobacter pylori. J. Bacteriol. 189, 8914–8921 (2007).
Bjorkholm, B. M. et al. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277, 34191–34197 (2002).
Kong, H. et al. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 28, 3216–3223 (2000).
Vitkute, J. et al. Specificities of eleven different DNA methyltransferases of Helicobacter pylori strain 26695. J. Bacteriol. 183, 443–450 (2001).
Brocchi, M., Vasconcelos, A. & Zaha, A. Restriction-modification systems in Mycoplasma spp. Genet. Mol. Biol. 30, 236–244 (2007).
Gumulak-Smith, J. et al. Variations in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol. 40, 1037–1044 (2001).
Rao, D. N., Saha, S. & Krishnamurthy, V. ATP-dependent restriction enzymes. Prog. Nucleic Acid Res. Mol. Biol. 64, 1–63 (2000).
Titheradge, A. J., King, J., Ryu, J. & Murray, N. E. Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res. 29, 4195–4205 (2001).
Chin, V., Valinluck, V., Magaki, S. & Ryu, J. KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system. Nucleic Acids Res. 32, e138 (2004).
Gough, J. A. & Murray, N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J. Mol. Biol. 166, 1–19 (1983).
Murray, N. E. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology 148, 3–20 (2002).
Titheradge, A. J., Ternent, D. & Murray, N. E. A third family of allelic hsd genes in Salmonella enterica: sequence comparisons with related proteins identify conserved regions implicated in restriction of DNA. Mol. Microbiol. 22, 437–447 (1996).
Davies, G. P., Kemp, P., Molineux, I. J. & Murray, N. E. The DNA translocation and ATPase activities of restriction-deficient mutants of EcoKI. J. Mol. Biol. 292, 787–796 (1999).
Ellis, D. J., Dryden, D. T., Berge, T., Edwardson, J. M. & Henderson, R. M. Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Nature Struct. Biol. 6, 15–17 (1999).
Goodsell, D. S. Inside a living cell. Trends Biochem. Sci. 16, 203–206 (1991).
Dreier, J., MacWilliams, M. P. & Bickle, T. A. DNA cleavage by the type IC restriction-modification enzyme EcoR124II. J. Mol. Biol. 264, 722–733 (1996).
Janscak, P., MacWilliams, M. P., Sandmeier, U., Nagaraja, V. & Bickle, T. A. DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J. 18, 2638–2647 (1999).
Szczelkun, M. D. Kinetic models of translocation, head-on collision, and DNA cleavage by type I restriction endonucleases. Biochemistry 41, 2067–2074 (2002).
Pingoud, A. & Jeltsch, A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29, 3705–3727 (2001). A review of the structure and function of type II R–M systems.
Saunders, N. J. et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37, 207–215 (2000).
Snyder, L. A., Butcher, S. A. & Saunders, N. J. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147, 2321–2332 (2001).
Mrazek, J., Spormann, A. M. & Karlin, S. Genomic comparisons among gamma-proteobacteria. Environ. Microbiol. 8, 273–288 (2006).
Acknowledgements
This work was supported by the National Health and Medical Research Council (Australia) Research Program (grants 565526 and 519704) and the Australian Research Council Discovery Projects scheme (grant DP0879604).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Rights and permissions
About this article
Cite this article
Srikhanta, Y., Fox, K. & Jennings, M. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol 8, 196–206 (2010). https://doi.org/10.1038/nrmicro2283
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2283
This article is cited by
-
Genome-wide methylome analysis of two strains belonging to the hypervirulent Neisseria meningitidis serogroup W ST-11 clonal complex
Scientific Reports (2021)
-
Complete genome and methylome analysis of Neisseria meningitidis associated with increased serogroup Y disease
Scientific Reports (2020)
-
The bacterial epigenome
Nature Reviews Microbiology (2020)
-
Genetic variation regulates the activation and specificity of Restriction-Modification systems in Neisseria gonorrhoeae
Scientific Reports (2019)
-
Deciphering bacterial epigenomes using modern sequencing technologies
Nature Reviews Genetics (2019)