Key Points
-
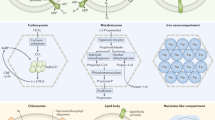
Planctomycetes form a distinctive phylum of the Bacteria, having a unique combination of features such as peptidoglycan-lacking proteinaceous cell walls, and intracellular membranes that form separate compartments within the cell cytoplasm. Such compartments are proving to be functionally as well as structurally distinct entities and are important for our understanding of the origin of the eukaryotic cell.
-
Planctomycetes are present in many different habitats, including the oceans, marine sediments, freshwater lakes, wastewater and terrestrial soils.
-
Anammox planctomycetes form a distinct family within the phylum and are anaerobic autotrophs that can oxidize ammonia to dinitrogen without oxygen. They play a major part in the global nitrogen cycle and form the basis for new industrial processes aimed at remediating nitrogen-rich wastewater. The unique physiology of these organisms is based on the presence of a specialized organelle, the anammoxosome, which might represent a bacterial analogue of the eukaryotic mitochondrion.
-
Comparative genomics and proteomics have revealed that the distinction between anammox and non-anammox planctomycetes goes beyond ammonium metabolism. Genomics has also reinforced the view that planctomycetes are members of a wider Planctomycetes–Verrucomicrobia–Chlamydiae (PVC) superphylum.
-
An endocytosis-like mechanism for the uptake of macromolecules such as proteins has been described in the planctomycete genus Gemmata. This mechanism is analogous, and possibly even homologous, to the eukaryotic process of endocytosis.
-
Research on planctomycetes is shedding new light on the origin and evolution of the eukaryotic endomembrane systems. In particular, it challenges current models for the origin of the nucleus that depend on the fusion of cells from different domains of life.
Abstract
Planctomycetes form a distinct phylum of the domain Bacteria and possess unusual features such as intracellular compartmentalization and a lack of peptidoglycan in their cell walls. Remarkably, cells of the genus Gemmata even contain a membrane-bound nucleoid analogous to the eukaryotic nucleus. Moreover, the so-called 'anammox' planctomycetes have a unique anaerobic, autotrophic metabolism that includes the ability to oxidize ammonium; this process is dependent on a characteristic membrane-bound cell compartment called the anammoxosome, which might be a functional analogue of the eukaryotic mitochondrion. The compartmentalization of planctomycetes challenges our hypotheses regarding the origins of eukaryotic organelles. Furthermore, the recent discovery of both an endocytosis-like ability and proteins homologous to eukaryotic clathrin in a planctomycete marks this phylum as one to watch for future research on the origin and evolution of the eukaryotic cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Buckley, D. H., Huangyutitham, V., Nelson, T. A., Rumberger, A. & Thies, J. E. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl. Environ. Microbiol. 72, 4522–4531 (2006).
Schlesner, H. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17, 135–145 (1994).
Fuerst, J. A. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141, 1493–1506 (1995).
Brummer, I. H., Felske, A. D. & Wagner-Dobler, I. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl. Environ. Microbiol. 70, 5094–5101 (2004).
Woebken, D. et al. Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 1, 419–435 (2007). Important comparative genomic analysis of all the available planctomycete genome data, revealing the absence of C 1 transfer genes in anammox organisms.
Goffredi, S. K. & Orphan, V. J. Bacterial community shifts in taxa and diversity in response to localized organic loading in the deep sea. Environ. Microbiol. 12, 344–363 (2010).
Drees, K. P. et al. Bacterial community structure in the hyperarid core of the Atacama Desert, Chile. Appl. Environ. Microbiol. 72, 7902–7908 (2006).
Kuypers, M. M. et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl Acad. Sci. USA 102, 6478–6483 (2005). First evidence of the global relevance of anammox planctomycetes for the nitrogen cycle.
Frey, J. C. et al. Fecal bacterial diversity in a wild gorilla. Appl. Environ. Microbiol. 72, 3788–3792 (2006).
Schabereiter-Gurter, C., Sar-Jiminez, C., Pinar, G., Lubitz, W. & Rolleke, S. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 211, 7–11 (2002).
Fukunaga, Y. et al. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275 (2009).
Cayrou, C., Raoult, D. & Drancourt, M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J. Antimicrob. Chemother. 65, 2119–2122 (2010).
Wang, J., Jenkins, C., Webb, R. I. & Fuerst, J. A. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 68, 417–422 (2002).
Kulichevskaya, I. S. et al. Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int. J. Syst. Evol. Microbiol. 59, 357–364 (2009).
Kulichevskaya, I. S. et al. Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 58, 1186–1193 (2008).
Kulichevskaya, I. S. et al. Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands. Int. J. Syst. Evol. Microbiol. 57, 2680–2687 (2007).
Winkelmann, N. & Harder, J. An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J. Microbiol. Methods 77, 276–284 (2009).
Bondoso, J. et al. Aquisphaera giovannonii gen. nov., sp. nov. A novel planctomycete isolated from a freshwater aquarium. Int. J. Syst. Evol. Microbiol. 14 Jan 2011 (doi:ijs.0.027474-0).
Kartal, B. et al. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30, 39–49 (2007).
Quan, Z. X. et al. Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ. Microbiol. 10, 3130–3139 (2008).
Schmid, M. et al. Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26, 529–538 (2003).
Kuenen, J. G. Anammox bacteria: from discovery to application. Nature Rev. Microbiol. 6, 320–326 (2008).
Pearson, A., Budin, M. & Brocks, J. J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl Acad. Sci. USA 100, 15352–15357 (2003).
Desmond, E. & Gribaldo, S. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1, 364–381 (2009).
Chistoserdova, L. et al. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21, 1234–1241 (2004).
Bauer, M. et al. Archaea-like genes for C1-transfer enzymes in Planctomycetes: phylogenetic implications of their unexpected presence in this phylum. J. Mol. Evol. 59, 571–586 (2004).
Woese, C. R. Bacterial evolution. Microbiol. Rev. 51, 221–271 (1987).
Strous, M. et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794 (2006). Genomic study of the anammox planctomycete ' Ca. Kuenenia stuttgartiensis' in relation to its physiology and the relationships between planctomycetes and chlamydiae.
Jun, S. R., Sims, G. E., Wu, G. A. & Kim, S. H. Whole-proteome phylogeny of prokaryotes by feature frequency profiles: An alignment-free method with optimal feature resolution. Proc. Natl Acad. Sci. USA 107, 133–138 (2010).
Cavalier-Smith, T. Deep phylogeny, ancestral groups and the four ages of life. Phil. Trans. R. Soc. B. 365, 111–132 (2010).
Cavalier-Smith, T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol. Direct 5, 7 (2010).
König, E., Schlesner, H. & Hirsch, P. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch. Microbiol. 138, 200–205 (1984).
Liesack, W., König, H., Schlesner, H. & Hirsch, P. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch. Microbiol. 145, 361–366 (1986).
Kerger, B. D. et al. The budding bacteria, Pirellula and Planctomyces, with atypical 16S rRNA and absence of peptidoglycan, show eubacterial phospholipids and uniquely high proportions of long chain beta-hydroxy fatty acids in the lipoplysaccharide lipid A. Arch. Microbiol. 149, 255–260 (1988).
Glockner, F. O. et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl Acad. Sci. USA 100, 8298–8303 (2003). Only published detailed account of the significant annotated features of a complete planctomycete genome.
Brochier, C. & Philippe, H. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature 417, 244 (2002).
Wagner, M. & Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249 (2006).
Glockner, J. et al. Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229 (2010).
Lee, K. C. et al. Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 9, 5 (2009). Demonstration that the planctomycete shared cell plan is also found in another bacterial phylum, the Verrucomicrobia.
Fieseler, L., Horn, M., Wagner, M. & Hentschel, U. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70, 3724–3732 (2004).
Siegl, A. et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70 (2010).
Wagar, E. A., Schachter, J., Bavoil, P. & Stephens, R. S. Differential human serologic response to two 60,000 molecular weight Chlamydia trachomatis antigens. J. Infect. Dis. 162, 922–927 (1990).
Papineau, D., Walker, J. J., Mojzsis, S. J. & Pace, N. R. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71, 4822–4832 (2005).
Kohler, T., Stingl, U., Meuser, K. & Brune, A. Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.). Environ. Microbiol. 10, 1260–1270 (2008).
Musat, N. et al. Microbial community structure of sandy intertidal sediments in the North Sea, Sylt-Romo Basin, Wadden Sea. Syst. Appl. Microbiol. 29, 333–348 (2006).
Chouari, R. et al. Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. Appl. Environ. Microbiol. 69, 7354–7363 (2003).
Fuerst, J. A. et al. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63, 254–262 (1997).
Gade, D. et al. Identification of planctomycetes with order-, genus-, and strain-specific 16S rRNA-targeted probes. Microb. Ecol. 47, 243–251 (2004).
Delong, E. F., Franks, D. G. & Alldredge, A. L. Phylogenetic diversity of aggregate-attached and free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934 (1993).
Morris, R. M., Longnecker, K. & Giovannoni, S. J. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8, 1361–1370 (2006).
Bengtsson, M. M. & Ovreas, L. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 10, 261 (2010).
Kuypers, M. M. et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422, 608–611 (2003).
Woebken, D. et al. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ. Microbiol. 10, 3106–3119 (2008).
Kirkpatrick, J. et al. Diversity and distribution of Planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol 72, 3079–3083 (2006).
Jetten, M. S. The microbial nitrogen cycle. Environ. Microbiol. 10, 2903–2909 (2008).
van der Star, W. R. et al. Startup of reactors for anoxic ammonium oxidation: experiences from the first full-scale anammox reactor in Rotterdam. Water Res. 41, 4149–4163 (2007).
Kartal, B., Kuenen, J. G. & van Loosdrecht, M. C. Engineering. Sewage treatment with anammox. Science 328, 702–703 (2010).
Giovannoni, S., Schabtach, E. & Castenholz, R. W. Isosphaera pallida gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch. Microbiol. 147, 276–284 (1987).
Pierson, B. K., Giovannoni, S. J., Stahl, D. A. & Castenholz, R. W. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch. Microbiol. 142, 164–167 (1985).
Li, H., Chen, S., Mu, B. Z. & Gu, J. D. Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb. Ecol. 60, 771–783 (2010).
Byrne, N. et al. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 3, 117–123 (2009).
Vergin, K. L. et al. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64, 3075–3078 (1998).
Shu, Q. & Jiao, N. New primers for amplification of the planctomycetes 16SrRNA gene from environmental samples. J. Rapid Methods Autom. Microbiol. 16, 330–336 (2008).
Lindsay, M. R., Webb, R. I. & Fuerst, J. A. Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology 143, 739–748 (1997). First recognition of a major intracellular membrane-bound compartment that is now known to be present in all planctomycetes.
Lindsay, M. R. et al. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175, 413–429 (2001).
Fuerst, J. A. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 59, 299–328 (2005). Review of planctomycete structure, cell plan and compartmentalization.
Butler, M. K., Wang, J., Webb, R. I. & Fuerst, J. A. Molecular and ultrastructural confirmation of classification of ATCC 35122 as a strain of Pirellula staleyi. Int. J. Syst. Evol. Microbiol. 52, 1663–1667 (2002).
van Niftrik, L. et al. Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium 'Candidatus Kuenenia stuttgartiensis'. Mol. Microbiol. 77, 701–715 (2010). Evidence that ATP synthase is found in membranes of the anammox planctomycete anammoxosome, consistent with a model for anammox biochemistry involving PMF generation.
Fuerst, J. A. & Webb, R. I. Membrane-bounded nucleoid in the eubacterium Gemmatata obscuriglobus. Proc. Natl Acad. Sci. USA 88, 8184–8188 (1991).
Lieber, A., Leis, A., Kushmaro, A., Minsky, A. & Medalia, O. Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus. J. Bacteriol. 191, 1439–1445 (2009).
Santarella-Mellwig, R. et al. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8, e1000281 (2010). Study establishing the presence of membrane coat-like proteins homologous to COP-family proteins and clathrins in planctomycetes, and the discovery of a homologue of a eukaryotic membrane coat-like protein in G. obscuriglobus.
Lee, K. C., Webb, R. I. & Fuerst, J. A. The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol. 10, 4 (2009).
Giovannoni, S. J., Godchaux, W. 3rd, Schabtach, E. & Castenholz, R. W. Cell wall and lipid composition of Isosphaera pallida, a budding eubacterium from hot springs. J. Bacteriol. 169, 2702–2707 (1987).
Hieu, C. X. et al. Detailed proteome analysis of growing cells of the planctomycete Rhodopirellula baltica SH1T. Proteomics 8, 1608–1623 (2008).
Jenkins, C. et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl Acad. Sci. USA 99, 17049–17054 (2002).
Pilhofer, M., Rosati, G., Ludwig, W., Schleifer, K. H. & Petroni, G. Coexistence of tubulins and ftsZ in different Prosthecobacter species. Mol. Biol. Evol. 24, 1439–1442 (2007).
Makarova, K. S. & Koonin, E. V. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol. Direct 5, 33 (2010).
Jetten, M. S. et al. Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44, 65–84 (2009).
van Niftrik, L. A. et al. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol. Lett. 233, 7–13 (2004).
Sinninghe Damste, J. S., Rijpstra, W. I., Geenevasen, J. A., Strous, M. & Jetten, M. S. Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox). FEBS J. 272, 4270–4283 (2005).
Sinninghe Damste, J. S. et al. A mixed ladderane/n-alkyl glycerol diether membrane lipid in an anaerobic ammonium-oxidizing bacterium. Chem. Commun. (Camb.) 2004, 2590–2591 (2004).
Sinninghe Damste, J. S. et al. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature 419, 708–712 (2002).
Boumann, H. A. et al. Biophysical properties of membrane lipids of anammox bacteria: I. Ladderane phospholipids form highly organized fluid membranes. Biochim. Biophys. Acta 1788, 1444–1451 (2009).
Karlsson, R., Karlsson, A., Backman, O., Johansson, B. R. & Hulth, S. Identification of key proteins involved in the anammox reaction. FEMS Microbiol. Lett. 297, 87–94 (2009).
Kartal, B., Keltjens, J. & Jetten, M. S. M. in Nitrification (eds. Ward, B. B., Arp, D. J. & Klotz, M. G.) (ASM Press, Washington DC, 2011).
van Niftrik, L. et al. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190, 708–717 (2008).
van der Star, W. R. et al. An intracellular pH gradient in the anammox bacterium Kuenenia stuttgartiensis as evaluated by 31P NMR. Appl. Microbiol. Biotechnol. 86, 311–317 (2010).
Medema, M. H. et al. A predicted physicochemically distinct sub-proteome associated with the intracellular organelle of the anammox bacterium Kuenenia stuttgartiensis. BMC Genomics 11, 299 (2010).
Belkin, S., Mehlhorn, R. J. & Packer, L. Proton gradients in intact cyanobacteria. Plant Physiol. 84, 25–30 (1987).
van Niftrik, L. et al. Cell division ring, a new cell division protein and vertical inheritance of a bacterial organelle in anammox planctomycetes. Mol. Microbiol. 73, 1009–1019 (2009).
Rachel, R. Another way to divide: the case of anammox bacteria. Mol. Microbiol. 73, 978–981 (2009).
Lindas, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J. & Bernander, R. A unique cell division machinery in the Archaea. Proc. Natl Acad. Sci. USA 105, 18942–18946 (2008).
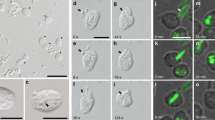
Lonhienne, T. G. et al. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl Acad. Sci. USA 107, 12883–12888 (2010). First description of macromolecule uptake in bacteria that is similar to receptor-mediated eukaryotic endocytosis.
Field, M. C. & Dacks, J. B. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 21, 4–13 (2009).
Dacks, J. B. & Field, M. C. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120, 2977–2985 (2007).
de Duve, C. The origin of eukaryotes: a reappraisal. Nature Rev. Genet. 8, 395–403 (2007).
Clum, A. et al. Complete genome sequence of Pirellula staleyi type strain (ATCC 27377T). Stand. Genomic Sci. 1, 308–316 (2009).
LaButti, K. et al. Complete genome sequence of Planctomyces limnophilus type strain (Mü 290T). Stand. Genomic Sci. 3, 47–56 (2010).
Gadler, P. & Faber, K. New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo- and enantioselectivity. Trends Biotechnol. 25, 83–88 (2007).
Gadler, P. et al. Biocatalytic approaches for the quantitative production of single stereoisomers from racemates. Biochem. Soc. Trans. 34, 296–300 (2006).
Wecker, P. et al. Life cycle analysis of the model orgnism Rhodopirellula baltica SH 1T by transcriptome studies. Microb. Biotechnol. 3, 583–594 (2010).
Lombardot, T., Bauer, M., Teeling, H., Amann, R. & Glockner, F. O. The transcriptional regulator pool of the marine bacterium Rhodopirellula baltica SH 1T as revealed by whole genome comparisons. FEMS Microbiol. Lett. 242, 137–145 (2005).
Gade, D. et al. Towards the proteome of the marine bacterium Rhodopirellula baltica: mapping the soluble proteins. Proteomics 5, 3654–3671 (2005).
Gade, D., Stuhrmann, T., Reinhardt, R. & Rabus, R. Growth phase dependent regulation of protein composition in Rhodopirellula baltica. Environ. Microbiol. 7, 1074–1084 (2005).
Gade, D., Gobom, J. & Rabus, R. Proteomic analysis of carbohydrate catabolism and regulation in the marine bacterium Rhodopirellula baltica. Proteomics 5, 3672–3683 (2005).
Lucker, S. et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl Acad. Sci. USA 107, 13479–13484 (2010).
Jenkins, C., Kedar, V. & Fuerst, J. A. Gene discovery within the planctomycete division of the domain Bacteria using sequence tags from genomic DNA libraries. Genome Biol. 3, RESEARCH0031 (2002).
Staley, J. T., Bouzek, H. & Jenkins, C. Eukaryotic signature proteins of Prosthecobacter dejongeii and Gemmata sp. Wa-1 as revealed by in silico analysis. FEMS Microbiol. Lett. 243, 9–14 (2005).
Forterre, P. & Gribaldo, S. Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proc. Natl Acad. Sci. USA 107, 12739–12740 (2010).
Poole, A. M. Did group II intron proliferation in an endosymbiont-bearing archaeon create eukaryotes? Biol. Direct 1, 36 (2006).
Lopez, D. & Kolter, R. Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902 (2010).
Tucker, J. D. et al. Membrane invagination in Rhodobacter sphaeroides is initiated at curved regions of the cytoplasmic membrane, then forms both budded and fully detached spherical vesicles. Mol. Microbiol. 76, 833–847 (2010).
Lopez, D., Vlamakis, H., Losick, R. & Kolter, R. Paracrine signaling in a bacterium. Genes Dev. 23, 1631–1638 (2009).
Fuerst, J. A. & Sagulenko, E. Protein uptake by bacteria: an endocytosis-like process in the planctomycete Gemmata obscuriglobus. Commun. Integr. Biol. 3, 572–575 (2010).
Acknowledgements
Research on planctomycetes in the J.A.F. laboratory is supported by the Australian Research Council (ARC Discovery Project DP0881485).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Nucleoid
-
The region of the bacterial cell that contains the genomic DNA, usually seen in thin sections as a fibrillar region; in electron micrographs of cryosubstituted Escherichia coli cell sections, the nucleoid seems to occupy much of the cell, whereas it forms a highly condensed fibrillar structure in planctomycetes.
- Anammox
-
Anaerobic ammonium oxidation, a process performed by some species of planctomycetes, whereby ammonium is oxidized to dinitrogen using nitrite as an electron acceptor via intermediates, including toxic hydrazine.
- C1 transfer
-
The process by which a compound containing only one carbon atom (a C1 compound; such as methane, methylamine, formate and formaldehyde) is enzymatically transformed into another C1 compound.
- Sulphatases
-
Enzymes that hydrolytically cleave sulphate esters to yield inorganic sulphate and an alcohol.
- Clathrin
-
A eukaryotic protein that coats the vesicles formed during endocytosis.
- Anammoxosome
-
An organelle within the pirellulosome of anammox planctomycetes that is surrounded by a single bilayer membrane and contains enzymes that are essential for the oxidation of ammonia to dinitrogen.
- Heterolactic fermentation
-
A pathway for anaerobic fermentation of carbohydrates; typically found in lactic acid bacteria, in which the dominant end products are lactic acid, ethanol and carbon dioxide.
- Divisome
-
A complex of proteins that is associated with cell division in peptidoglycan-synthesizing bacteria and that locates to the septum of cells dividing by binary fission. The complex includes cell division protein FtsZ and, usually, other proteins such as FtsI, FtsA and FtsK, as well as penicillin-binding proteins involved in peptidoglycan synthesis.
Rights and permissions
About this article
Cite this article
Fuerst, J., Sagulenko, E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9, 403–413 (2011). https://doi.org/10.1038/nrmicro2578
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2578