Key Points
-
Tailed bacteriophages package their double-stranded DNA chromosomes into preformed protein containers called procapsids.
-
The DNA-packaging process condenses the DNA several-hundred-fold and requires chemical energy. The result of this condensation is DNA that is arranged as a tightly wound, imperfect solenoid that is held within the virion with no proteins holding the DNA strands close together.
-
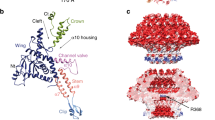
The molecular motor that packages DNA within the procapsid is a DNA translocase that is composed of three proteins: portal protein, which forms the hole through which DNA enters the procapsid, and also senses when the capsid is full of DNA; small terminase subunit (TerS), which recognizes the DNA that will be packaged; and large terminase subunit (TerL), the ATPase that converts the chemical energy of ATP hydrolysis into physical motion of the DNA.
-
In the phages that package over-length or circular DNA, TerL also contains a nuclease activity that cuts the DNA to form the unit-length, linear DNA molecules that are found in completed virions.
-
Single-molecule experiments have shown that the DNA-packaging motor can insert DNA into the procapsid at rates of up to ∼1,800 bp per second, generates forces of up to 100 piconewtons, and translocates the DNA in steps of 10 bp that in turn consist of four rapid 2.5-bp substeps.
-
TerL and probably TerS are physical components of the packaging motor that are released from the nascent virion upon completion of packaging, so they are not present in completed virions.
-
After DNA packaging and TerL release, other proteins stabilize the packaged DNA by plugging the channel through which the DNA entered the procapsid and by binding the outside of the capsid shell to strengthen it.
-
Recently obtained atomic structures of packaging motor proteins, channel plug proteins and shell-strengthening proteins, as well as subnanometer-resolution, cryo-electron microscopy-derived three-dimensional reconstructions of virions and procapsids, have shed new light on the mechanism of the motor and its control.
Abstract
Tailed bacteriophages use nanomotors, or molecular machines that convert chemical energy into physical movement of molecules, to insert their double-stranded DNA genomes into virus particles. These viral nanomotors are powered by ATP hydrolysis and pump the DNA into a preformed protein container called a procapsid. As a result, the virions contain very highly compacted chromosomes. Here, I review recent progress in obtaining structural information for virions, procapsids and the individual motor protein components, and discuss single-molecule in vitro packaging reactions, which have yielded important new information about the mechanism by which these powerful molecular machines translocate DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bergh, O., Borsheim, K. Y., Bratbak, G. & Heldal, M. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (1989).
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005).
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Wommack, K. E. & Colwell, R. R. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114 (2000).
Brussow, H. & Hendrix, R. W. Phage genomics: small is beautiful. Cell 108, 13–16 (2002).
Boyd, E. F., Davis, B. M. & Hochhut, B. Bacteriophage–bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9, 137–144 (2001).
Casjens, S. & Hendrix, R. in The Bacterial Chromosome (ed. Higgins, N. P.) 39–52 (American Society for Microbiology Press, Washingtion DC, 2005).
Cheetham, B. F. & Katz, M. E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18, 201–208 (1995).
Denou, E. et al. T4 phages against Escherichia coli diarrhea: potential and problems. Virology 388, 21–30 (2009).
Summers, W. C. Bacteriophage therapy. Annu. Rev. Microbiol. 55, 437–451 (2001).
Luftig, R. B., Wood, W. B. & Okinaka, R. Bacteriophage T4 head morphogenesis. On the nature of gene 49-defective heads and their role as intermediates. J. Mol. Biol. 57, 555–573 (1971).
Kaiser, D., Syvanen, M. & Masuda, T. DNA packaging steps in bacteriophage lambda head assembly. J. Mol. Biol. 91, 175–186 (1975).
Catalano, C. (ed.) Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (Landes Bioscience, Georgetown, Texas, 2005).
Casjens, S. & Hendrix, R. in The Bacteriophages (ed. Calendar, R.) 15–91 (Plenum, New York, 1988).
Earnshaw, W. & Casjens, S. DNA packaging by the double-stranded DNA bacteriophages. Cell 21, 319–331 (1980).
Black, L. W. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43, 267–292 (1989).
Maluf, N. K. & Feiss, M. Virus DNA translocation: progress towards a first ascent of Mount Pretty Difficult. Mol. Microbiol. 61, 1–4 (2006).
Rao, V. B. & Feiss, M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42, 647–681 (2008).
Jeembaeva, M., Jonsson, B., Castelnovo, M. & Evilevitch, A. DNA heats up: energetics of genome ejection from phage revealed by isothermal titration calorimetry. J. Mol. Biol. 395, 1079–1087 (2010).
Panja, D. & Molineux, I. J. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys. Biol. 7, 045006 (2010).
Kindt, J., Tzlil, S., Ben-Shaul, A. & Gelbart, W. M. DNA packaging and ejection forces in bacteriophage. Proc. Natl Acad. Sci. USA 98, 13671–13674 (2001).
Riemer, S. C. & Bloomfield, V. A. Packaging of DNA in bacteriophage heads: some considerations on energetics. Biopolymers 17, 785–794 (1978).
Agirrezabala, X. et al. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 24, 3820–3829 (2005).
Choi, K. H. et al. Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J. Mol. Biol. 378, 726–736 (2008).
Jiang, W. et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439, 612–616 (2006).
Lander, G. C. et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006).
Lander, G. C. et al. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16, 1399–1406 (2008). This report describes the subnanometre, cryo-electron microscopy-derived three-dimensional reconstruction of phage λ capsids, with a focus on the bacteriophage shell-strengthening protein.
Leiman, P. G. et al. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 371, 836–849 (2007).
Tang, J. et al. DNA poised for release in bacteriophage φ29. Structure 16, 935–943 (2008).
Chang, J., Weigele, P., King, J., Chiu, W. & Jiang, W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14, 1073–1082 (2006).
Duda, R. L., Hendrix, R. W., Huang, W. M. & Conway, J. F. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr. Biol. 16, R11–R13 (2006).
Liu, X. et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nature Struct. Mol. Biol. 17, 830–836 (2010).
Fokine, A. et al. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl Acad. Sci. USA 101, 6003–6008 (2004).
Effantin, G., Boulanger, P., Neumann, E., Letellier, L. & Conway, J. F. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 361, 993–1002 (2006).
Dai, W. et al. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl Acad. Sci. USA 107, 4347–4352 (2010).
Cerritelli, M. E. et al. Encapsidated conformation of bacteriophage T7 DNA. Cell 91, 271–280 (1997).
Leforestier, A. & Livolant, F. Structure of toroidal DNA collapsed inside the phage capsid. Proc. Natl Acad. Sci. USA 106, 9157–9162 (2009).
Agirrezabala, X. et al. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347, 895–902 (2005).
Cerritelli, M. E. et al. A second symmetry mismatch at the portal vertex of bacteriophage T7, 8-fold symmetry in the procapsid core. J. Mol. Biol. 327, 1–6 (2003).
Tang, J. et al. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure 19, 496–502 (2011).
Black, L. W. & Silverman, D. J. Model for DNA packaging into bacteriophage T4 heads. J. Virol. 28, 643–655 (1978).
de Frutos, M., Letellier, L. & Raspaud, E. DNA ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys. J. 88, 1364–1370 (2005).
Leforestier, A. & Livolant, F. The bacteriophage genome undergoes a succession of intracapsid phase transitions upon DNA ejection. J. Mol. Biol. 396, 384–395 (2010).
Sao-Jose, C., de Frutos, M., Raspaud, E., Santos, M. A. & Tavares, P. Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol. 374, 346–355 (2007).
Letellier, L., Boulanger, P., Plancon, L., Jacquot, P. & Santamaria, M. Main features on tailed phage, host recognition and DNA uptake. Front. Biosci. 9, 1228–1339 (2004).
Casjens, S. & Molineux, I. in Viral Molecular Machines (eds Rossmann, M. & Rao, V.) (Springer, New York, in the press).
Casjens, S. R. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8, 451–458 (2005).
Hendrix, R. W. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61, 471–480 (2002).
Kochan, J., Carrascosa, J. L. & Murialdo, H. Bacteriophage lambda preconnectors: purification and structure. J. Mol. Biol. 174, 433–447 (1984).
Driedonks, R. A., Engel, A., tenHeggeler, B. & van Driel, R. Gene 20 product of bacteriophage T4 its purification and structure. J. Mol. Biol. 152, 641–662 (1981).
Carazo, J. M., Donate, L. E., Herranz, L., Secilla, J. P. & Carrascosa, J. L. Three-dimensional reconstruction of the connector of bacteriophage φ29 at 1.8 nm resolution. J. Mol. Biol. 192, 853–867 (1986).
Lurz, R. et al. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310, 1027–1037 (2001).
Doan, D. N. & Dokland, T. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J. Struct. Biol. 157, 432–436 (2007).
Morais, M. C. et al. Defining molecular and domain boundaries in the bacteriophage φ29 DNA packaging motor. Structure 16, 1267–1274 (2008). This investigation uses cryo-electron microscopy to obtain a three-dimensional reconstruction of the phage φ29 procapsid with the TerL ATPase bound to it.
Simpson, A. A. et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408, 745–750 (2000).
Lebedev, A. A. et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 26, 1984–1994 (2007).
Olia, A., Prevelige, P. E. Jr, Johnson, J. & Cingolani, G. Three-dimensional structure of a viral genome-delivery portal vertex. Nature Struc. Mol. Biol. 18, 597–604 (2011). This article describes the most recent of the three X-ray structures of portal protein rings (see also references 55 and 56).
Zheng, H. et al. A conformational switch in bacteriophage P22 portal protein primes genome injection. Mol. Cell 29, 376–383 (2008).
Cuervo, A., Vaney, M. C., Antson, A. A., Tavares, P. & Oliveira, L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J. Biol. Chem. 282, 18907–18913 (2007).
Chen, D. H. et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl Acad. Sci. USA 108, 1355–1360 (2011). This paper describes the highest-resolution asymmetrical three-dimensional reconstruction of procapsids that has been obtained to date by cryo-electron microscopy.
Bazinet, C. & King, J. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202, 77–86 (1988).
Weigele, P., Sampson, L., Winn-Stapley, D. & Casjens, S. Molecular genetics of bacteriophage P22 scaffolding protein's functional domains. J. Mol. Biol. 348, 831–844 (2005).
Hendrix, R. W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl Acad. Sci. USA 75, 4779–4783 (1978).
Baumann, R. G., Mullaney, J. & Black, L. W. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 61, 16–32 (2006).
Hugel, T. et al. Experimental test of connector rotation during DNA packaging into bacteriophage φ29 capsids. PLoS Biol. 5, e59 (2007).
Ding, F. et al. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc. Natl Acad. Sci. USA 108, 7357–7362 (2011).
Reid, R., Zhang, F., Benson, S. & Anderson, D. Probing the structure of bacteriophage φ29 prohead RNA with specific mutations. J. Biol. Chem. 269, 18656–18661 (1994).
Zhang, C., Tellinghuisen, T. & Guo, P. Confirmation of the helical structure of the 5′/3′ termini of the essential DNA packaging pRNA of phage φ29. RNA 1, 1041–1050 (1995).
Casjens, S. & Gilcrease, D. in Bacteriophages: Methods and Protocols (eds Clokie, M. & Kropinski, A.) 91–111 (Humana, Totowa, New Jersey, 2009).
Jackson, E. N., Jackson, D. A. & Deans, R. J. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118, 365–388 (1978).
Tye, B. K., Huberman, J. A. & Botstein, D. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 85, 501–528 (1974).
Adams, M. B., Hayden, M. & Casjens, S. On the sequential packaging of bacteriophage P22 DNA. J. Virol. 46, 673–677 (1983).
Meijer, W. J., Horcajadas, J. A. & Salas, M. φ29 family of phages. Microbiol. Mol. Biol. Rev. 65, 261–287 (2001).
Mousset, S. & Thomas, R. Ter, a function which generates the ends of the mature λ chromosome. Nature 221, 242–245 (1969).
Poteete, A. R. & Botstein, D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology 95, 565–573 (1979).
Al-Zahrani, A. S. et al. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J. Biol. Chem. 284, 24490–24500 (2009).
Nemecek, D. et al. Subunit conformations and assembly states of a DNA-translocating motor: the terminase of bacteriophage P22. J. Mol. Biol. 374, 817–836 (2007).
Gual, A., Camacho, A. G. & Alonso, J. C. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 275, 35311–35319 (2000).
Leffers, G. & Rao, V. B. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 275, 37127–37136 (2000).
Maluf, N. K., Yang, Q. & Catalano, C. E. Self-association properties of the bacteriophage l terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 347, 523–542 (2005).
Sun, S. et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262 (2008). This study uses cryo-electron microscopy to obtain a three-dimensionsal reconstruction of the phage T4 procapsid with the TerL ATPase bound to it.
Burroughs, A. M., Iyer, L. M. & Aravind, L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 3, 48–65 (2007).
Mitchell, M. S., Matsuzaki, S., Imai, S. & Rao, V. B. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30, 4009–4021 (2002).
Guo, P., Peterson, C. & Anderson, D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J. Mol. Biol. 197, 229–236 (1987).
Hwang, Y. & Feiss, M. Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage l terminase, inactivate the endonuclease activity of terminase. J. Mol. Biol. 261, 524–535 (1996).
Morita, M., Tasaka, M. & Fujisawa, H. DNA packaging ATPase of bacteriophage T3. Virology 193, 748–752 (1993).
Oliveira, L., Henriques, A. O. & Tavares, P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 281, 21914–21923 (2006).
Goetzinger, K. R. & Rao, V. B. Defining the ATPase center of bacteriophage T4 DNA packaging machine: requirement for a catalytic glutamate residue in the large terminase protein gp17. J. Mol. Biol. 331, 139–154 (2003).
Duffy, C. & Feiss, M. The large subunit of bacteriophage l's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 316, 547–561 (2002).
Kondabagil, K. R., Zhang, Z. & Rao, V. B. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 363, 786–799 (2006).
Tsay, J. M. et al. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 285, 24282–24289 (2010).
Tsay, J. M., Sippy, J., Feiss, M. & Smith, D. E. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl Acad. Sci. USA 106, 14355–14360 (2009).
Alam, T. I. et al. The headful packaging nuclease of bacteriophage T4. Mol. Microbiol. 69, 1180–1190 (2008).
Smits, C. et al. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 10, 592–598 (2009).
Hwang, Y., Hang, J. Q., Neagle, J., Duffy, C. & Feiss, M. Endonuclease and helicase activities of bacteriophage l terminase: changing nearby residue 515 restores activity to the gpA K497D mutant enzyme. Virology 277, 204–214 (2000).
Zhang, Z. et al. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 9, e1000592 (2011).
Oliveira, L., Alonso, J. C. & Tavares, P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. J. Mol. Biol. 353, 529–539 (2005).
de Beer, T. et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 9, 981–991 (2002).
Hanagan, A., Meyer, J. D., Johnson, L., Manning, M. C. & Catalano, C. E. The phage lambda terminase enzyme: 2. Refolding of the gpNu1 subunit from the detergent-denatured and guanidinium hydrochloride-denatured state yields different oligomerization states and altered protein stabilities. Int. J. Biol. Macromol. 23, 37–48 (1998).
Nemecek, D., Lander, G. C., Johnson, J. E., Casjens, S. R. & Thomas, G. J. Jr. Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J. Mol. Biol. 383, 494–501 (2008).
Zhao, H. et al. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl Acad. Sci. USA 107, 1971–1976 (2010). This report presents the only currently known X-ray structure of TerS, the protein that is responsible for recognition of the DNA that will be packaged.
Lin, H., Simon, M. N. & Black, L. W. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 272, 3495–3501 (1997).
Chai, S., Lurz, R. & Alonso, J. C. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 252, 386–398 (1995).
Casjens, S. R. & Thuman-Commike, P. A. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411, 393–415 (2011).
Casjens, S. in Virus Structure and Assembly (ed. Casjens, S.) 75–148 (Jones and Bartlett, Boston, 1985).
Sun, S., Rao, V. B. & Rossmann, M. G. Genome packaging in viruses. Curr. Opin. Struct. Biol. 20, 114–120 (2010).
Serwer, P. et al. DNA packaging-associated hyper-capsid expansion of bacteriophage T3. J. Mol. Biol. 397, 361–374 (2010).
Yu, T. Y. & Schaefer, J. REDOR NMR characterization of DNA packaging in bacteriophage T4. J. Mol. Biol. 382, 1031–1042 (2008).
Koti, J. S. et al. DNA packaging motor assembly intermediate of bacteriophage φ29. J. Mol. Biol. 381, 1114–1132 (2008).
Maluf, N. K., Gaussier, H., Bogner, E., Feiss, M. & Catalano, C. E. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry 45, 15259–15268 (2006).
Chemla, Y. R. et al. Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 (2005).
Smith, D. et al. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001). This important paper describes the first application of single-molecule optical tweezer technology to phage DNA packaging. It describes the basic rate and force properties of the phage ϕ29 motor.
Gope, R. & Serwer, P. Bacteriophage P22 in vitro DNA packaging monitored by agarose gel electrophoresis: rate of DNA entry into capsids. J. Virol. 47, 96–105 (1983).
Yang, Q., Catalano, C. E. & Maluf, N. K. Kinetic analysis of the genome packaging reaction in bacteriophage l. Biochemistry 48, 10705–10715 (2009).
Laemmli, U. K. & Favre, M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80, 575–599 (1973).
Fuller, D. N. et al. Measurements of single DNA molecule packaging dynamics in bacteriophage l reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 373, 1113–1122 (2007). This study defines the basic rate and force parameters of the phage λ packaging motor.
Fuller, D. N., Raymer, D. M., Kottadiel, V. I., Rao, V. B. & Smith, D. E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl Acad. Sci. USA 104, 16868–16873 (2007). This investigation defines the basic rate and force parameters of the phage T4 packaging motor.
Rickgauer, J. P. et al. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage φ29. Biophys. J. 94, 159–167 (2008).
Moffitt, J. R. et al. Intersubunit coordination in a homomeric ring ATPase. Nature 457, 446–450 (2009). This important work refines optical tweezer analysis to be able to study the step size of DNA packaging (10 bp) and the coordination of the 2.5-bp substeps.
Yu, J., Moffitt, J., Hetherington, C. L., Bustamante, C. & Oster, G. Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 400, 186–203 (2010).
Draper, B. & Rao, V. B. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 369, 79–94 (2007).
Ray, K., Sabanayagam, C. R., Lakowicz, J. R. & Black, L. W. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology 398, 224–232 (2010).
Ortega, M. E., Gaussier, H. & Catalano, C. E. The DNA maturation domain of gpA, the DNA packaging motor protein of bacteriophage lambda, contains an ATPase site associated with endonuclease activity. J. Mol. Biol. 373, 851–865 (2007).
Ghosh-Kumar, M., Alam, T. I., Draper, B., Stack, J. D. & Rao, V. B. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 39, 2742–2755 (2011).
Yang, Q. & Catalano, C. E. A minimal kinetic model for a viral DNA packaging machine. Biochemistry 43, 289–299 (2004).
Gao, S. & Rao, V. B. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J. Biol. Chem. 286, 3944–3956 (2011).
Woods, L., Terpening, C. & Catalano, C. E. Kinetic analysis of the endonuclease activity of phage l terminase: assembly of a catalytically competent nicking complex is rate-limiting. Biochemistry 36, 5777–5785 (1997).
Gaussier, H., Ortega, M. E., Maluf, N. K. & Catalano, C. E. Nucleotides regulate the conformational state of the small terminase subunit from bacteriophage lambda: implications for the assembly of a viral genome-packaging motor. Biochemistry 44, 9645–9656 (2005).
Aathavan, K. et al. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461, 669–673 (2009).
Oram, M., Sabanayagam, C. & Black, L. W. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 381, 61–72 (2008).
Leffers, G. & Rao, V. B. A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J. Mol. Biol. 258, 839–850 (1996).
Xin, W. & Feiss, M. Function of IHF in l DNA packaging: I. Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J. Mol. Biol. 230, 492–504 (1993).
Wu, H., Sampson, L., Parr, R. & Casjens, S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45, 1631–1646 (2002).
Bukhari, A. I., Froshauer, S. & Botchan, M. Ends of bacteriophage Mu DNA. Nature 264, 580–583 (1976).
Groenen, M. A. & van de Putte, P. Mapping of a site for packaging of bacteriophage Mu DNA. Virology 144, 520–522 (1985).
Harel, J., Duplessis, L., Kahn, J. S. & DuBow, M. S. The cis-acting DNA sequences required in vivo for bacteriophage Mu helper-mediated transposition and packaging. Arch. Microbiol. 154, 67–72 (1990).
Casjens, S. et al. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339, 379–394 (2004).
Casjens, S. R. et al. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187, 1091–1104 (2005).
Black, L. W. & Peng, G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J. Biol. Chem. 281, 25635–25643 (2006).
Zhang, X. & Studier, F. W. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J. Mol. Biol. 340, 707–730 (2004).
Chung, Y. B., Nardone, C. & Hinkle, D. C. Bacteriophage T7 DNA packaging. III. A “hairpin” end formed on T7 concatemers may be an intermediate in the processing reaction. J. Mol. Biol. 216, 939–948 (1990).
Stewart, C. R. et al. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388, 48–70 (2009).
Xin, W., Cai, Z. H. & Feiss, M. Function of IHF in l DNA packaging: II. Effects of mutations altering the IHF binding site and the intrinsic bend in cosB on l development. J. Mol. Biol. 230, 505–515 (1993).
Droge, A. & Tavares, P. in Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (ed. Catalano, C.) 89–101 (Landes Bioscience, Georgetown, Texas, 2005).
Feiss, M. & Catalano, C. in Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (ed. Catalano, C.) 5–39 (Landes Bioscience, Georgetown, Texas, 2005).
Frackman, S., Siegele, D. A. & Feiss, M. A functional domain of bacteriophage l terminase for prohead binding. J. Mol. Biol. 180, 283–300 (1984).
Morita, M., Tasaka, M. & Fujisawa, H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 245, 635–644 (1995).
Oliveira, L., Cuervo, A. & Tavares, P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J. Biol. Chem. 285, 7366–7373 (2010).
Lin, H., Rao, V. B. & Black, L. W. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 289, 249–260 (1999).
Casjens, S. et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 224, 1055–1074 (1992).
Isidro, A., Santos, M. A., Henriques, A. O. & Tavares, P. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol. Microbiol. 51, 949–962 (2004).
Wieczorek, D. J. & Feiss, M. Defining cosQ, the site required for termination of bacteriophage l DNA packaging. Genetics 158, 495–506 (2001).
Cardarelli, L. et al. The crystal structure of bacteriophage HK97 gp6: defining a large family of head–tail connector proteins. J. Mol. Biol. 395, 754–768 (2010).
Lhuillier, S. et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl Acad. Sci. USA 106, 8507–8512 (2009).
Olia, A. S. et al. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol. 363, 558–576 (2006).
Olia, A. S., Bhardwaj, A., Joss, L., Casjens, S. & Cingolani, G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry 46, 8776–8784 (2007).
Olia, A. S., Casjens, S. & Cingolani, G. Structure of phage P22 cell envelope-penetrating needle. Nature Struct. Mol. Biol. 14, 1221–1227 (2007).
Chattoraj, D. K. & Inman, R. B. Location of DNA ends in P2, 186, P4 and lambda bacteriophage heads. J. Mol. Biol. 87, 11–22 (1974).
Saigo, K. & Uchida, H. Connection of the right-hand terminus of DNA to the proximal end of the tail in bacteriophage lambda. Virology 61, 524–536 (1974).
Thomas, J. O., Sternberg, N. & Weisberg, R. Altered arrangement of the DNA in injection-defective lambda bacteriophage. J. Mol. Biol. 123, 149–161 (1978).
Plisson, C. et al. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26, 3720–3728 (2007).
Israel, V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J. Virol. 23, 91–97 (1977).
Wikoff, W. R. et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 (2000).
Gilcrease, E. B., Winn-Stapley, D. A., Hewitt, F. C., Joss, L. & Casjens, S. R. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol. 187, 2050–2057 (2005).
Tang, L., Gilcrease, E. B., Casjens, S. R. & Johnson, J. E. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure 14, 837–845 (2006).
Qin, L., Fokine, A., O'Donnell, E., Rao, V. B. & Rossmann, M. G. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J. Mol. Biol. 395, 728–741 (2010).
Sathaliyawala, T. et al. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 77, 444–455 (2010).
Yang, Q., Maluf, N. K. & Catalano, C. E. Packaging of a unit-length viral genome: the role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage l. J. Mol. Biol. 383, 1037–1048 (2008).
Lee, T. J., Schwartz, C. & Guo, P. Construction of bacteriophage φ29 DNA packaging motor and its applications in nanotechnology and therapy. Ann. Biomed. Eng. 37, 2064–2081 (2009).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nature Biotech. 26, 1146–1153 (2008).
Sun, S., Kondabagil, K., Gentz, P. M., Rossmann, M. G. & Rao, V. B. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell 25, 943–949 (2007).
Casjens, S. in Structural Biology of Viruses (eds Chiu, W., Burnett, R. & Garcea, R.) 3–37 (Oxford Univ. Press, Oxford, 1997).
Fuller, D. N. et al. Ionic effects on viral DNA packaging and portal motor function in bacteriophage ϕ29. Proc. Natl Acad. Sci. USA 104, 11245–11250 (2007). This paper describes the X-ray structure of phage T4 TerL, the ATPase that powers DNA packaging.
Acknowledgements
I thank M. Feiss, V. Rao and S. Grimes for thoughtful reading of this manuscript before publication. My research is supported by the US National Institutes of Health grant RO1 AI074825.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- Tailed bacteriophages
-
Double-stranded-DNA bacteriophages with a protein tail that attaches to a susceptible bacterium. DNA is injected into the host through the tail.
- Prophages
-
Bacteriophage genomes that are physically integrated into the chromosome of the host bacterium.
- Nanomotor
-
A molecular motor that functions on a nanometre scale to convert energy into directed physical movement of molecules.
- Procapsid
-
The preformed protein container into which DNA is packaged during virion assembly.
- Portal protein
-
The part of the DNA-packaging nanomotor that forms the hole or portal through the phage capsid; DNA enters and exits the virion through this portal.
- Portal vertex
-
The unique vertex to which the portal protein and tails are attached. Icosahedral structures such as phage heads have 12 five-fold rotationally symmetrical vertices.
- Low-angle X-ray scattering
-
A technique that is used to determine the radially averaged electron density of particles that make up an unoriented (as opposed to crystalline) sample. It is based on the deflection of a beam of X-rays away from its straight trajectory after it interacts with the particles in the sample.
- Terminase
-
The DNA nanomotor protein complex that recognizes the DNA which will be packaged. The terminase contains the ATPase activity that converts chemical energy into mechanical movement, and also contains the active site for cleavage of double-stranded DNA (when such a site is present).
- Concatemers
-
Long DNA molecules that contain multiple head-to-tail repeats of the phage genome sequence. Concatemers are often formed as the result of the DNA synthesis machinery replicating around a circular template multiple times without terminating.
- Occam's razor
-
A line of reasoning that argues that the simplest explanation should be favoured until a more complex one is required by the observed data (attributed to the Franciscan friar Father William of Ockham).
- Late-gene transcription factors
-
The phage-encoded early proteins (which are expressed early during infection) that cause transcription of the genes which are expressed at late times after infection.
- IHF
-
(Integration host factor). An Escherichia coli protein that binds DNA and bends it at a sharp angle. IHF was first discovered as a cofactor of the phage λ enzyme integrase.
Rights and permissions
About this article
Cite this article
Casjens, S. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol 9, 647–657 (2011). https://doi.org/10.1038/nrmicro2632
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2632
This article is cited by
-
Design of bacteriophage T4-based artificial viral vectors for human genome remodeling
Nature Communications (2023)
-
Isolation, Characterization, and Comparative Genomic Analysis of vB_Pd_C23, a Novel Bacteriophage of Pantoea dispersa
Current Microbiology (2023)
-
Characterization and genome analysis of Escherichia phage fBC-Eco01, isolated from wastewater in Tunisia
Archives of Virology (2023)
-
Whole genome sequence analysis of bacteriophage P1 that infects the Lactobacillus plantarum
Virus Genes (2022)
-
A viral genome packaging ring-ATPase is a flexibly coordinated pentamer
Nature Communications (2021)