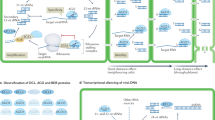
Key Points
-
Coevolution between plants and pathogens has given rise to a large variety of defence pathways and counter-acting virulence pathways. Plant resistance against bacterial and fungal pathogens is largely mediated by a first layer of basal resistance and a second layer of strong race-specific resistance; successful pathogens have evolved effector molecules to suppress both layers of defence. Analogously, virus resistance is mediated broadly by RNA silencing and also by race-specific resistance.
-
Viruses actively suppress RNA silencing, which alleviates anti-viral defence but also disrupts endogenous host gene regulation through microRNAs and small-interfering RNAs. Virulent infections thus cause disease partly through disrupting host silencing, while viruses unable to suppress silencing fail to replicate in healthy plants.
-
RNA silencing actively contributes to defence against bacterial and eukaryotic pathogens as well, through regulation of host gene expression. These non-viral pathogens have consequently evolved silencing suppressors that contribute to disease.
-
Recent work has revealed a crucial role for multiple aspects of host silencing in repressing race-specific defence responses. Thus, resistance and defence-promoting genes are silenced by miRNAs, siRNAs and/or epigenetically through DNA methylation.
-
Silencing suppressors deployed by pathogens therefore prevent the first layer of defence responses, but relieve repression of strong race-specific resistance mechanisms, which can contribute to increased host resistance.
-
This review synthesizes the intricate interplay between plant resistance pathways, RNA silencing and its manipulation by pathogens.
Abstract
RNA silencing is a central regulator of gene expression in most eukaryotes and acts both at the transcriptional level through DNA methylation and at the post-transcriptional level through direct mRNA interference mediated by small RNAs. In plants and invertebrates, the same pathways also function directly in host defence against viruses by targeting viral RNA for degradation. Successful viruses have consequently evolved diverse mechanisms to avoid silencing, most notably through the expression of viral suppressors of RNA silencing. RNA silencing suppressors have also been recently identified in plant pathogenic bacteria and oomycetes, suggesting that disruption of host silencing is a general virulence strategy across several kingdoms of plant pathogens. There is also increasing evidence that plants have evolved specific defences against RNA-silencing suppression by pathogens, providing yet another illustration of the never-ending molecular arms race between plant pathogens and their hosts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Brodersen, P. & Voinnet, O. The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280 (2006).
Sunkar, R., Li, Y. F. & Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203 (2012).
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010).
Yu, A. et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl Acad. Sci. USA 110, 2389–2394 (2013).
Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 (2009).
Ding, S. W. & Voinnet, O. Antiviral immunity directed by small RNAs. Cell 130, 413–426 (2007).
Zipfel, C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004).
Shan, L. et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27 (2008).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Zhai, J. et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553 (2011).
Li, F. et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl Acad. Sci. USA 109, 1790–1795 (2012).
Shivaprasad, P. V. et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874 (2012). References 10, 11 and 12 uncovered a role for siRNAs in repressing plant resistance genes
Navarro, L., Jay, F., Nomura, K., He, S. Y. & Voinnet, O. Suppression of the microRNA pathway by bacterial effector proteins. Science 321, 964–967 (2008). First report of a silencing suppressor outside of viruses.
Qiao, Y. et al. Oomycete pathogens encode RNA silencing suppressors. Nature Genet. 45, 330–333 (2013). First discovery of a silencing suppressor in a eukaryotic pathogen.
Ruiz-Ferrer, V. & Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510 (2009).
Pagan, I. et al. Arabidopsis thaliana as a model for the study of plant-virus co-evolution. Phil. Trans. R. Soc. B 365, 1983–1995 (2010).
Burgyan, J. & Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272 (2011).
Qu, F., Ye, X. & Morris, T. J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl Acad. Sci. USA 105, 14732–14737 (2008).
Jakubiec, A., Yang, S. W. & Chua, N. H. Arabidopsis DRB4 protein in antiviral defense against Turnip yellow mosaic virus infection. Plant J. 69, 14–25 (2012).
Deleris, A. et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006).
Garcia-Ruiz, H. et al. Arabidopsis RNA-dependent RNA polymerases and Dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22, 481–496 (2010).
Wang, X. B. et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell 23, 1625–1638 (2011).
Vogler, H. et al. Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J. Virol. 81, 10379–10388 (2007).
Morel, J. B. et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639 (2002).
Zhang, X., Zhang, X., Singh, J., Li, D. & Qu, F. Temperature-dependent survival of turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires DCL2, AGO2, and HEN1. J. Virol. 86, 6847–6854 (2012).
Harvey, J. J. et al. An antiviral defense role of AGO2 in plants. PLoS ONE 6, e14639 (2011).
Jaubert, M. J., Bhattacharjee, S., Mello, A. F., Perry, K. L. & Moffett, P. ARGONAUTE2 mediates RNA silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564 (2011).
Scholthof, H. B. et al. Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 156, 1548–1555 (2011).
Azevedo, J. et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24, 904–915 (2010). References 21, 22 and 29 detail genetic work with subtle viral mutations that disrupt silencing suppressors.
Akbergenov, R. et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471 (2006).
Carbonell, A. et al. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24, 3613–3629 (2012).
Vaucheret, H. Plant ARGONAUTES. Trends Plant Sci. 13, 350–358 (2008).
Laliberte, J. F. & Sanfacon, H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48, 69–91 (2010).
Brodersen, P. et al. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 1778–1783 (2012).
Li, S. et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574 (2013). References 34 and 35 reveal a functional association between AGO1 and membranes.
Brodersen, P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008).
Lanet, E. et al. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21, 1762–1768 (2009).
Miozzi, L., Gambino, G., Burgyan, J. & Pantaleo, V. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 14, 30–43 (2013).
Pantaleo, V., Szittya, G. & Burgyan, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 81, 3797–3806 (2007).
Hoffer, P. et al. Posttranscriptional gene silencing in nuclei. Proc. Natl Acad. Sci. USA 108, 409–414 (2011).
Kumakura, N. et al. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583, 1261–1266 (2009).
Mourrain, P. et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542 (2000).
Schwach, F., Vaistij, F. E., Jones, L. & Baulcombe, D. C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus x and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852 (2005).
Gy, I. et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007).
Gazzani, S., Lawerson, T., Woodward, D., Headon, R. & Sablowski, R. A link between mRNA turnover and RNA interference in Arabidopsis. Science 306, 1046–1048 (2004).
Moreno, A. B. et al. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 41, 4699–4708 (2013).
Diaz-Pendon, J. A., Li, F., Li, W. X. & Ding, S. W. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19, 2053–2063 (2007).
Donaire, L. et al. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82, 5167–5177 (2008).
Wang, X. B. et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 107, 484–489 (2010).
Qi, X., Bao, F. S. & Xie, Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4, e4971 (2009).
Dunoyer, P. et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science 328, 912–916 (2010).
Molnar, A. et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 (2011).
Montgomery, T. A. et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141 (2008).
Mi, S. et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008).
Brigneti, G. et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746 (1998).
Anandalakshmi, R. et al. A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA 95, 13079–13084 (1998).
Kasschau, K. D. & Carrington, J. C. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95, 461–470 (1998).
Voinnet, O., Pinto, Y. M. & Baulcombe, D. C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl Acad. Sci. USA 96, 14147–14152 (1999).
Schott, G. et al. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 31, 2553–2565 (2012).
Baumberger, N., Tsai, C. H., Lie, M., Havecker, E. & Baulcombe, D. C. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614 (2007).
Csorba, T., Lozsa, R., Hutvagner, G. & Burgyan, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472 (2010).
Derrien, B. et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl Acad. Sci. USA 109, 15942–15946 (2012).
Gibbings, D. et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nature Cell Biol. 14, 1314–1321 (2012).
Zhang, P. & Zhang, H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 14, 568–576 (2013).
Chiu, M. H., Chen, I. H., Baulcombe, D. C. & Tsai, C. H. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11, 641–649 (2010).
Duan, C. G. et al. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24, 259–274 (2012).
Haas, G. et al. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 27, 2102–2112 (2008).
Guo, H. et al. Rice yellow stunt rhabdovirus protein 6 suppresses systemic RNA silencing by blocking RDR6-mediated secondary siRNA synthesis. Mol. Plant Microbe Interact. 26, 927–936 (2013).
Glick, E. et al. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl Acad. Sci. USA 105, 157–161 (2008).
Cuellar, W. J. et al. Elimination of antiviral defense by viral RNase III. Proc. Natl Acad. Sci. USA 106, 10354–10358 (2009).
Ye, K., Malinina, L. & Patel, D. J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426, 874–878 (2003).
Vargason, J. M., Szittya, G., Burgyan, J. & Tanaka Hall, T. M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115, 799–811 (2003).
Varallyay, E., Valoczi, A., Agyi, A., Burgyan, J. & Havelda, Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519 (2010).
Mallory, A. C. & Vaucheret, H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10, 521–526 (2009).
Mallory, A. C. et al. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 5, e1000646 (2009).
Varallyay, E. & Havelda, Z. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 14, 567–575 (2013).
Wu, Q. et al. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl Acad. Sci. USA 109, 3938–3943 (2012).
Flores, R. et al. Viroids: the minimal non-coding RNAs with autonomous replication. FEBS Lett. 567, 42–48 (2004).
Itaya, A. et al. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81, 2980–2994 (2007).
Denti, M. A., Boutla, A., Tsagris, M. & Tabler, M. Short interfering RNAs specific for potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J. 37, 762–769 (2004).
Blevins, T. et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39, 5003–5014 (2011).
Moissiard, G. & Voinnet, O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl Acad. Sci. USA 103, 19593–19598 (2006).
Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 (2003).
Zhang, X. et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268 (2006).
Chapman, E. J., Prokhnevsky, A. I., Gopinath, K., Dolja, V. V. & Carrington, J. C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186 (2004).
Dunoyer, P., Lecellier, C. H., Parizotto, E. A., Himber, C. & Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16, 1235–1250 (2004).
Lichner, Z., Silhavy, D. & Burgyan, J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84, 975–980 (2003).
Bucher, E., Hemmes, H., de Haan, P., Goldbach, R. & Prins, M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85, 983–991 (2004).
Himber, C., Dunoyer, P., Moissiard, G., Ritzenthaler, C. & Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22, 4523–4533 (2003).
Havelda, Z., Hornyik, C., Crescenzi, A. & Burgyan, J. In situ characterization of Cymbidium ringspot Tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77, 6082–6086 (2003).
Chen, H. Y., Yang, J., Lin, C. & Yuan, Y. A. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760 (2008).
Bayne, E. H., Rakitina, D. V., Morozov, S. Y. & Baulcombe, D. C. Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482 (2005).
Lakatos, L. et al. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780 (2006).
Fahlgren, N. et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22, 1074–1089 (2010).
Cuperus, J. T., Fahlgren, N. & Carrington, J. C. Evolution and functional diversification of MIRNA genes. Plant Cell 23, 431–442 (2011).
Navarro, L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006).
Katiyar-Agarwal, S. & Jin, H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246 (2010).
Fahlgren, N. et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2, e219 (2007).
Xin, M. et al. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10, 123 (2010).
Lu, S., Sun, Y. H., Amerson, H. & Chiang, V. L. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 51, 1077–1098 (2007).
Guo, N. et al. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 54, 954–958 (2011).
Li, Y. et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152, 2222–2231 (2010).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005).
Fu, J. & Wang, S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2, 74 (2011).
Zhang, X. et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366 (2011).
Ellendorff, U., Fradin, E. F., de Jonge, R. & Thomma, B. P. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602 (2009).
Katiyar-Agarwal, S. et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl Acad. Sci. USA 103, 18002–18007 (2006).
Katiyar-Agarwal, S., Gao, S., Vivian-Smith, A. & Jin, H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21, 3123–3134 (2007).
Wang, X. J., Gaasterland, T. & Chua, N. H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 6, R30 (2005).
Raja, P., Wolf, J. N. & Bisaro, D. M. RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799, 337–351 (2010).
Chellappan, P., Vanitharani, R. & Fauquet, C. M. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 78, 7465–7477 (2004).
Raja, P., Sanville, B. C., Buchmann, R. C. & Bisaro, D. M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007 (2008).
Dogar, A. M. RNAi dependent epigenetic marks on a geminivirus promoter. Virol. J. 3, 5 (2006).
Al-Kaff, N. S. et al. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279, 2113–2115 (1998).
Seemanpillai, M., Dry, I., Randles, J. & Rezaian, A. Transcriptional silencing of geminiviral promoter-driven transgenes following homologous virus infection. Mol. Plant Microbe Interact. 16, 429–438 (2003).
Yang, X. et al. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7, e1002329 (2011).
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G. & Bisaro, D. M. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013 (2009).
Wang, H., Buckley, K. J., Yang, X., Buchmann, R. C. & Bisaro, D. M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79, 7410–7418 (2005).
Zhang, Z. et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23, 273–288 (2011).
Rodriguez-Negrete, E. et al. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475 (2013).
Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M. & Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77 (2003).
Pavet, V., Quintero, C., Cecchini, N. M., Rosa, A. L. & Alvarez, M. E. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by pseudomonas syringae. Mol. Plant Microbe Interact. 19, 577–587 (2006).
Dowen, R. H. et al. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl Acad. Sci. USA 109, E2183–E2191 (2012). With Reference 4, first demonstration that RdDM represses plant defence.
Lopez, A., Ramirez, V., Garcia-Andrade, J., Flors, V. & Vera, P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 7, e1002434 (2011).
Henderson, I. R. et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genet. 38, 721–725 (2006).
Kallman, T., Chen, J., Gyllenstrand, N. & Lagercrantz, U. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162, 741–754 (2013).
Palma, K. et al. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6, e1001137 (2010).
Bendahmane, A., Kanyuka, K. & Baulcombe, D. C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792 (1999).
Yi, H. & Richards, E. J. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19, 2929–2939 (2007).
Whitham, S. et al. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115 (1994).
Burch-Smith, T. M. et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5, e68 (2007).
Padmanabhan, M. S. et al. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 9, e1003235 (2013).
Li, H. W. et al. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18, 2683–2691 (1999).
Sansregret, R. et al. Extreme resistance as a host counter-counter defense against viral suppression of RNA silencing. PLoS Pathog. 9, e1003435 (2013). Demonstrates a strong defence response triggered specifically by the activity of a VSR.
Cooley, M. B., Pathirana, S., Wu, H. J., Kachroo, P. & Klessig, D. F. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676 (2000).
Wang, L. Y. et al. Multiple domains of the tobacco mosaic virus p126 protein can independently suppress local and systemic RNA silencing. Mol. Plant Microbe Interact. 25, 648–657 (2012).
Smith, N. A., Eamens, A. L. & Wang, M. B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7, e1002022 (2011). Clear demonstration of disease symptoms caused by a vsiRNA targeting a host gene.
Rasmann, S. et al. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158, 854–863 (2012).
Slaughter, A. et al. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158, 835–843 (2012).
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V. & Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853 (2012).
Kanazawa, A. et al. Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J. 65, 156–168 (2011).
Voinnet, O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13, 317–328 (2008).
Liu, Q., Feng, Y. & Zhu, Z. Dicer-like (DCL) proteins in plants. Funct. Integr. Genom. 9, 277–286 (2009).
Boutet, S. et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848 (2003).
Yu, B. et al. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 (2005).
Law, J. A. & Jacobsen, S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev. Genet. 11, 204–220 (2010).
Han, M. H., Goud, S., Song, L. & Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl Acad. Sci. USA 101, 1093–1098 (2004).
Zilberman, D., Cao, X. & Jacobsen, S. E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 (2003).
Kanno, T. et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805 (2004).
Zheng, X., Zhu, J., Kapoor, A. & Zhu, J. K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26, 1691–1701 (2007).
Matzke, M., Kanno, T., Daxinger, L., Huettel, B. & Matzke, A. J. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376 (2009).
Chen, H. M. et al. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl Acad. Sci. USA 107, 15269–15274 (2010).
Cuperus, J. T. et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Struct. Mol. Biol. 17, 997–1003 (2010).
Manavella, P. A., Koenig, D. & Weigel, D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl Acad. Sci. USA 109, 2461–2466 (2012).
Gomez-Gomez, L. & Boller, T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 (2000).
Chinchilla, D. et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 (2007).
van der Hoorn, R. A. & Kamoun, S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017 (2008).
Friedman, A. R. & Baker, B. J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 17, 493–499 (2007).
Belkhadir, Y., Nimchuk, Z., Hubert, D. A., Mackey, D. & Dangl, J. L. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16, 2822–2835 (2004).
Lorrain, S., Vailleau, F., Balague, C. & Roby, D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271 (2003).
Lecellier, C. H. et al. A cellular microRNA mediates antiviral defense in human cells. Science 308, 557–560 (2005).
Lu, S. & Cullen, B. R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 78, 12868–12876 (2004).
Cullen, B. R. MicroRNAs as mediators of viral evasion of the immune system. Nature Immunol. 14, 205–210 (2013).
Umbach, J. L. et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783 (2008).
Sullivan, C. S., Grundhoff, A. T., Tevethia, S., Pipas, J. M. & Ganem, D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435, 682–686 (2005).
Choy, E. Y. et al. An Epstein–Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205, 2551–2560 (2008).
Abend, J. R., Uldrick, T. & Ziegelbauer, J. M. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol. 84, 12139–12151 (2010).
Lewsey, M., Robertson, F. C., Canto, T., Palukaitis, P. & Carr, J. P. Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 50, 240–252 (2007).
Chen, J., Li, W. X., Xie, D., Peng, J. R. & Ding, S. W. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell 16, 1302–1313 (2004).
Jay, F. et al. Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 7, e1002035 (2011).
Wang, M. B. et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl Acad. Sci. USA 101, 3275–3280 (2004).
Zhu, S. et al. Double-stranded RNA-binding protein 4 is required for resistance signaling against viral and bacterial pathogens. Cell Rep. 4, 1168–1184 (2013)
Acknowledgements
Work in the O.V. group is supported by a core grant from ETH-Z, Advanced Researcher grant 'Frontiers of RNAi-II' (No. 323071), from the European Research Council, and a grant from the Swiss National Foundation (No. 31003A_132907). N.P. received support from an EMBO Long-Term Fellowship and a Marie-Curie Actions Incoming International Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- Pathogen-associated molecular patterns
-
(PAMPs; alternatively, microorganism-associated molecular patterns). Conserved and essential pathogen signature molecules that include flagellin, lipopoly-saccharide, elongation factors and double-stranded RNA.
- Oomycetes
-
Eukaryotic fungal-like organisms of the kingdom Stramenopila that include the causal agent of the Irish potato famine, Phytophthora infestans.
- Viral suppressors of RNA silencing
-
(VSRs). Proteins encoded by viruses that function as virulence factors by inhibiting post-transcriptional gene silencing pathways in plants and invertebrates.
- MicroRNA
-
(miRNA). Discrete 21- or 22-nucleotide small RNAs, arising from genome-encoded imperfect foldback structures processed by DCL1, which negatively regulate endogenous target mRNAs by complementary base-pairing.
- Repeat-associated small interfering RNA
-
(rasiRNA). 24-nucleotide small interfering RNAs generated from genomic repeats and transposon loci by DCL3, which target DNA methylation and chromatin modification at their loci of origin.
- Trans-acting siRNA
-
(tasiRNA). 21-nucleotide small interfering RNAs produced by sequential processing by DCL4 on long, perfectly complementary double-stranded RNA.
- hrcC
-
A mutant of Pseudomonas syringae pv. tomato that lacks a functional type III secretion system, thus making it unable to deliver virulence effectors into its host.
- Type III secretion system
-
(T3SS). A delivery system encoded by a wide range of Gram-negative bacterial pathogens of plants and animals that is used to inject effector proteins into host cells.
- Avirulent
-
A pathogen that elicits a specific effector-triggered immunity response owing to recognition of one of its virulence factors by a cognate host-encoded resistance gene, resulting in an incompatible interaction.
- SNARE
-
Soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein (SNAP) receptor, which regulates secretory vesicle fusion.
- PATHOGENESIS-RELATED 1
-
(PR1). A secreted protein with suspected antimicrobial functions. It is used as a classic defence marker owing to its strong induction during pathogen challenge.
- Hypersensitive response
-
(HR). A specific form of programmed cell death, often induced by effector-triggered immunity and correlated with accumulation of antimicrobial compounds and systemic acquired resistance.
- Bacterial suppressors of RNA silencing
-
(BSRs). Effector molecules used by bacterial pathogens to alter host post-transcriptional gene silencing or, possibly, transcriptional gene silencing.
- Chromocentres
-
Densely packed heterochromatic (silenced) DNA including pericentromeric regions, transposons and repetitive elements.
- Salicylic acid
-
A central plant hormone signal that induces local and systemic defence responses, including systemic acquired resistance.
- Biotrophic
-
Organisms that complete their life cycle on living plant tissue; such organisms actively prevent host cell death.
- Necrotrophic
-
Organisms that kill host cells before invasion and gain nutrients from the dead plant tissue.
- Jasmonic acid
-
A phytohormone that regulates defence against necrotrophic pathogens and herbivores.
- Helitron
-
A DNA transposon that is commonly subjected to repressive DNA methylation and that transposes through rolling circle DNA replication.
- Extreme resistance
-
A plant defence response against viruses that is elicited by effector-triggered immunity. It differs from the hypersensitive response in that it restricts viral accumulation without cell death.
Rights and permissions
About this article
Cite this article
Pumplin, N., Voinnet, O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11, 745–760 (2013). https://doi.org/10.1038/nrmicro3120
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3120
This article is cited by
-
Epigenetics: Toward improving crop disease resistance and agronomic characteristics
Plant Biotechnology Reports (2024)
-
P4 protein of an Indian isolate of rice tungro bacilliform virus modulates gene silencing
Virus Genes (2024)
-
Synergistic crop virus disease complexes in Sub-saharan Africa: causes, consequences and control
Phytoparasitica (2024)
-
Transcriptional and epigenetic changes during tomato yellow leaf curl virus infection in tomato
BMC Plant Biology (2023)
-
Identification of ceRNA-vsiRNA-mRNA network for exploring the mechanism underlying pathogenesis of sugarcane mosaic virus in resistant and susceptible maize inbred lines
Phytopathology Research (2023)