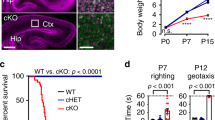
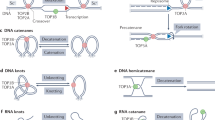
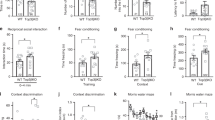
Abstract
Topoisomerases are unique enzymes that regulate torsional stress in DNA to enable essential genome functions, including DNA replication and transcription. Although all cells in an organism require topoisomerases to maintain normal function, the nervous system in particular shows a vital need for these enzymes. Indeed, a range of inherited human neurologic syndromes, including neurodegeneration, schizophrenia and intellectual impairment, are associated with aberrant topoisomerase function. Much remains unknown regarding the tissue-specific function of neural topoisomerases or the connections between these enzymes and disease aetiology. Precisely how topoisomerases regulate genome dynamics within the nervous system is therefore a crucial research question.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gartenberg, M. R. & Wang, J. C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl Acad. Sci. USA 89, 11461–11465 (1992).
Joshi, R. S., Pina, B. & Roca, J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. EMBO J. 29, 740–748 (2010).
Bermejo, R., Lai, M. S. & Foiani, M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell 45, 710–718 (2012).
McClendon, A. K., Rodriguez, A. C. & Osheroff, N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345 (2005).
Vos, S. M., Tretter, E. M., Schmidt, B. H. & Berger, J. M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 (2011).
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002).
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001).
Nitiss, J. L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 (2009).
Pommier, Y. et al. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid. Res. Mol. Biol. 81, 179–229 (2006).
Mondal, N. & Parvin, J. D. DNA topoisomerase IIα is required for RNA polymerase II transcription on chromatin templates. Nature 413, 435–438 (2001).
Pedersen, J. M. et al. DNA topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 8, e1003128 (2012).
Zhang, H., Wang, J. C. & Liu, L. F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl Acad. Sci. USA 85, 1060–1064 (1988).
Andersen, S. L., Sloan, R. S., Petes, T. D. & Jinks-Robertson, S. Genome-destabilizing effects associated with Top1 loss or accumulation of Top1 cleavage complexes in yeast. PLoS Genet. 11, e1005098 (2015).
Chen, S. H., Chan, N. L. & Hsieh, T. S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013).
Takashima, H. et al. Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272 (2002).
McKinnon, P. J. Maintaining genome stability in the nervous system. Nat. Neurosci. 16, 1523–1529 (2013).
Gómez-Herreros, F. et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat. Genet. 46, 516–521 (2014).
Stoll, G. et al. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237 (2013).
Xu, D. et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 16, 1238–1247 (2013).
Akimitsu, N. et al. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIα. Genes Cells 8, 393–402 (2003).
Li, W. & Wang, J. C. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc. Natl Acad. Sci. USA 95, 1010–1013 (1998).
Morham, S. G., Kluckman, K. D., Voulomanos, N. & Smithies, O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 16, 6804–6809 (1996).
King, I. F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013).
Lyu, Y. L. et al. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell. Biol. 26, 7929–7941 (2006).
Madabhushi, R. et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 (2015).
Keeney, S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2, 81–123 (2008).
Douarre, C. et al. Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS ONE 7, e41094 (2012).
Zhang, H. et al. Human mitochondrial topoisomerase I. Proc. Natl Acad. Sci. USA 98, 10608–10613 (2001).
Linka, R. M. et al. C-Terminal regions of topoisomerase IIα and IIβ determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35, 3810–3822 (2007).
Zhang, H. et al. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Res. 42, 7259–7267 (2014).
Wang, Y., Lyu, Y. L. & Wang, J. C. Dual localization of human DNA topoisomerase IIIα to mitochondria and nucleus. Proc. Natl Acad. Sci. USA 99, 12114–12119 (2002).
Yang, X., Li, W., Prescott, E. D., Burden, S. J. & Wang, J. C. DNA topoisomerase IIβ and neural development. Science 287, 131–134 (2000).
Kwan, K. Y. & Wang, J. C. Mice lacking DNA topoisomerase IIIβ develop to maturity but show a reduced mean lifespan. Proc. Natl Acad. Sci. USA 98, 5717–5721 (2001).
Khiati, S. et al. Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc. Natl Acad. Sci. USA 112, 11282–11287 (2015).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Lin, C. P., Ban, Y., Lyu, Y. L., Desai, S. D. & Liu, L. F. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J. Biol. Chem. 283, 21074–21083 (2008).
Mao, Y., Sun, M., Desai, S. D. & Liu, L. F. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl Acad. Sci. USA 97, 4046–4051 (2000).
Ashour, M. E., Atteya, R. & El-Khamisy, S. F. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer 15, 137–151 (2015).
Caldecott, K. W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 (2008).
Pommier, Y. et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19, 114–129 (2014).
Gomez-Herreros, F. et al. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 9, e1003226 (2013).
Nitiss, J. L. & Nitiss, K. C. Tdp2: a means to fixing the ends. PLoS Genet. 9, e1003370 (2013).
El-Khamisy, S. F. et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005).
Cortes Ledesma, F., El Khamisy, S. F., Zuma, M. C., Osborn, K. & Caldecott, K. W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678 (2009).
McKinnon, P. J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 10, 100–112 (2009).
Zylka, M. J., Simon, J. M. & Philpot, B. D. Gene length matters in neurons. Neuron 86, 353–355 (2015).
Tsutsui, K., Tsutsui, K., Sano, K., Kikuchi, A. & Tokunaga, A. Involvement of DNA topoisomerase IIβ in neuronal differentiation. J. Biol. Chem. 276, 5769–5778 (2001).
Lyu, Y. L. & Wang, J. C. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl Acad. Sci. USA 100, 7123–7128 (2003).
Mabb, A. M. et al. Topoisomerase 1 regulates gene expression in neurons through cleavage complex-dependent and -independent mechanisms. PLoS ONE 11, e0156439 (2016).
Plaschkes, I., Silverman, F. W. & Priel, E. DNA topoisomerase I in the mouse central nervous system: age and sex dependence. J. Comp. Neurol. 493, 357–369 (2005).
Katyal, S. et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat. Neurosci. 17, 813–821 (2014).
Zehorai, E., Eitan, E., Hershfinkel, M., Sekler, I. & Priel, E. Glutamate regulates the activity of topoisomerase I in mouse cerebellum. Mol. Neurobiol. 38, 242–252 (2008).
Tiwari, V. K. et al. Target genes of topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl Acad. Sci. USA 109, E934–E943 (2012).
Li, Y. et al. Topoisomerase IIβ is required for proper retinal development and survival of postmitotic cells. Biol. Open 3, 172–184 (2014).
Bunch, H. et al. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 6, 10191 (2015).
Ju, B. G. et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Puc, J. et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 160, 367–380 (2015).
Mabb, A. M. et al. Topoisomerase 1 inhibition reversibly impairs synaptic function. Proc. Natl Acad. Sci. USA 111, 17290–17295 (2014).
Gabel, H. W. et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015).
Sugino, K. et al. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 34, 12877–12883 (2014).
Huang, H. S. et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481, 185–189 (2012).
Powell, W. T. et al. R-Loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl Acad. Sci. USA 110, 13938–13943 (2013).
McKinnon, P. J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 7, 303–321 (2012).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Lasda, E. & Parker, R. Circular RNAs: diversity of form and function. RNA 20, 1829–1842 (2014).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Yang, Y. et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 (2014).
Bhakar, A. L., Dolen, G. & Bear, M. F. The pathophysiology of fragile X (and what it teaches us about synapses). Annu. Rev. Neurosci. 35, 417–443 (2012).
Yang, Y. et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023 (2010).
Seki, T., Seki, M., Onodera, R., Katada, T. & Enomoto, T. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIβ) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J. Biol. Chem. 273, 28553–28556 (1998).
Aguilera, A. & Garcia-Muse, T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012).
Skourti-Stathaki, K. & Proudfoot, N. J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396 (2014).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
Rialdi, A. et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 352, aad7993 (2016).
Acknowledgements
The author thanks the US National Institutes of Health (NIH) (NS-37956, CA-21765), the Cancer Center Support Grant (CCSG) (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St Jude Children's Research Hospital for financial support. Space constraints limited the number of primary research papers cited.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
DATABASES
Rights and permissions
About this article
Cite this article
McKinnon, P. Topoisomerases and the regulation of neural function. Nat Rev Neurosci 17, 673–679 (2016). https://doi.org/10.1038/nrn.2016.101
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn.2016.101
This article is cited by
-
Flaxseed oil fraction reverses cardiac remodeling at a molecular level: improves cardiac function, decreases apoptosis, and suppresses miRNA-29b and miRNA 1 gene expression
BMC Complementary Medicine and Therapies (2024)
-
ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes
Nature Communications (2023)
-
Defective repair of topoisomerase I induced chromosomal damage in Huntington’s disease
Cellular and Molecular Life Sciences (2022)
-
Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology
Nature Neuroscience (2021)
-
PARylation prevents the proteasomal degradation of topoisomerase I DNA-protein crosslinks and induces their deubiquitylation
Nature Communications (2021)