Key Points
-
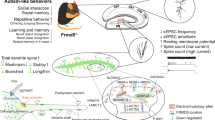
SH3 and multiple ankyrin repeat domains proteins (SHANKs) are encoded by SHANK1, SHANK2 and SHANK3 genes. The three different SHANK genes can produce multiple protein isoforms that are differentially expressed according to developmental stages, cell types and brain regions.
-
Mutations in SHANK genes are a potential monogenic cause for autism spectrum disorder.
-
Neurobiological studies in animal models indicate a wide array of functions for SHANK proteins, from synaptic scaffolding to regulating spine morphology and neurotransmission.
-
Mutant mice carrying different Shank1, Shank2 or Shank3 mutations have some distinct and shared phenotypes at the molecular and functional level. All mutants seem to have altered molecular composition of excitatory synapses and altered neurotransmission, and often display impaired social interaction and repetitive behaviour.
-
Different mutations within the same SHANKgene may cause distinct synaptic and circuitry defects and thus may be responsible for the different clinical features that are seen in patients.
-
Despite being a neurodevelopmental disorder, some neurobiological alterations in autism spectrum disorder may be reversible in adulthood.
-
Adult restoration of SHANK3 levels or restoration of downstream mediators may be a useful therapeutic approach to alleviate some of the synaptic and behavioural impairments that are associated with SHANK3 mutations.
Abstract
Several large-scale genomic studies have supported an association between cases of autism spectrum disorder and mutations in the genes SH3 and multiple ankyrin repeat domains protein 1 (SHANK1), SHANK2 and SHANK3, which encode a family of postsynaptic scaffolding proteins that are present at glutamatergic synapses in the CNS. An evaluation of human genetic data, as well as of in vitro and in vivo animal model data, may allow us to understand how disruption of SHANK scaffolding proteins affects the structure and function of neural circuits and alters behaviour.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sheng, M. & Hoogenraad, C. C. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847 (2007).
Ahmari, S. E. & Smith, S. J. Knowing a nascent synapse when you see it. Neuron 34, 333–336 (2002).
Kano, M. & Hashimoto, K. Synapse elimination in the central nervous system. Curr. Opin. Neurobiol. 19, 154–161 (2009).
Sheng, M. & Kim, E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 3, a005678 (2011).
Leblond, C. S. et al. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 10, e1004580 (2014). This study demonstrates a correlation between SHANK1, SHANK2 and SHANK3 mutations and the degree of cognitive impairment. The authors show that patients with SHANK3 mutations have more-severe cognitive deficits than those with SHANK1 or SHANK2 mutations and suggest SHANK mutation screening in clinical practice.
Gauthier, J. et al. Novel de novo SHANK3 mutation in autistic patients. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150, 421–424 (2009).
Boccuto, L. et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur. J. Hum. Genet. 21, 310–316 (2013).
Sala, C., Vicidomini, C., Bigi, I., Mossa, A. & Verpelli, C. Shank synaptic scaffold proteins: keys to understanding the pathogenesis of autism and other synaptic disorders. J. Neurochem. 135, 849–858 (2015).
Peça, J. et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011). This study describes the first Shank3 -mutant mice with PDZ domain deletion (exon 13 and exon 16 deletion). These mice show synaptic dysfunction and autism-related behavioural phenotypes such as impaired social interaction and stereotyped and/or repetitive behaviour.
Naisbitt, S. et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23, 569–582 (1999). This study provides the first description of the SHANK family of synaptic proteins.
Wang, X., Xu, Q., Bey, A. L., Lee, Y. & Jiang, Y. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Mol. Autism 5, 30 (2014). This study addresses the functional and transcriptional regulation of different SHANK3 isoforms. The results suggest that different SHANK3 isoforms have distinct functions and that the different SHANK3 mutations found in patients with ASD disrupt only specific isoforms and result in distinct phenotypes.
Jiang, Y. -H & Ehlers, M. D. Modeling autism by SHANK gene mutations in mice. Neuron 78, 8–27 (2013).
Beri, S. et al. DNA methylation regulates tissue-specific expression of Shank3. J. Neurochem. 101, 1380–1391 (2007).
Ching, T.-T. et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat. Genet. 37, 645–651 (2005).
Maunakea, A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Lim, S. et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 17, 385–397 (2001).
Böckers, T. M. et al. Synaptic scaffolding proteins in rat brain: ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein α-fodrin. J. Biol. Chem. 276, 40104–40112 (2001).
Mameza, M. G. et al. SHANK3 gene mutations associated with autism facilitate ligand binding to the shank3 ankyrin repeat region. J. Biol. Chem. 288, 26697–26708 (2013).
Uchino, S. et al. Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J. Neurochem. 97, 1203–1214 (2006).
Tu, J. C. et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23, 583–592 (1999).
Hayashi, M. K. et al. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 137, 159–171 (2009).
Baron, M. K. et al. An architectural framework that may lie at the core of the postsynaptic density. Science 311, 531–535 (2006).
Boeckers, T. M. et al. C-Terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J. Neurochem. 92, 519–524 (2005).
Grabrucker, A. M. et al. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 30, 569–581 (2011).
Guilmatre, A., Huguet, G., Delorme, R. & Bourgeron, T. The emerging role of SHANK genes in neuropsychiatric disorders. Dev. Neurobiol. 74, 113–122 (2014).
Boeckers, T. M., Bockmann, J., Kreutz, M. R. & Gundelfinger, E. D. ProSAP/Shank proteins — a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J. Neurochem. 81, 903–910 (2002).
Okamoto, P. M., Gamby, C., Wells, D., Fallon, J. & Vallee, R. B. Dynamin isoform-specific interaction with the Shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton. J. Biol. Chem. 276, 48458–48465 (2001).
McWilliams, R. R., Gidey, E., Fouassier, L., Weed, S. A. & Doctor, R. B. Characterization of an ankyrin repeat-containing Shank2 isoform (Shank2E) in liver epithelial cells. Biochem. J. 380, 181–191 (2004).
Leblond, C. S. et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 8, e1002521 (2012).
Schmeisser, M. J. et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 486, 256–260 (2012). This study describes one of the first Shank2 -mutant mice (exon 17 deletion; PDZ domain deletion). These mice show autism-related behavioural phenotypes and hyperactivity.
Lim, S. et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J. Biol. Chem. 274, 29510–29518 (1999).
Peça, J., Ting, J. & Feng, G. SnapShot: autism and the synapse. Cell 147, 706–706.e1 (2011).
Böckers, T. M. et al. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol. Cell. Neurosci. 26, 182–190 (2004).
Zitzer, H., Hönck, H. H., Bächner, D., Richter, D. & Kreienkamp, H. J. Somatostatin receptor interacting protein defines a novel family of multidomain proteins present in human and rodent brain. J. Biol. Chem. 274, 32997–33001 (1999).
Lee, J. et al. Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front. Cell. Neurosci. 9, 94 (2015).
Wang, X. et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 7, 11459 (2016).
Mei, Y. et al. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 530, 481–484 (2016). This is the first report to show that adult restoration of the Shank3 gene selectively rescues certain synaptic defects and some autism-related behavioural phenotypes such as social interaction and stereotyped and/or repetitive behaviour in mice.
Lai, M. C. et al. Cognition in males and females with autism: similarities and differences. PLoS ONE 7, e47198 (2012).
Centers for Disease Control and Prevention. Prevalence of autism spectrum disorders — Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill. Summ. 61, 1–19 (2012).
American Psychiatric Association (eds). Diagnostic and Statistical Manual of Mental Disorders 5th edn (American Psychiatric Association Publishing, 2013).
Rosenberg, R. E. et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatr. Adolesc. Med. 163, 907–914 (2009).
Geschwind, D. H. Advances in autism. Annu. Rev. Med. 60, 367–380 (2009).
Huguet, G., Ey, E. & Bourgeron, T. The genetic landscapes of autism spectrum disorders. Annu. Rev. Genomics Hum. Genet. 14, 191–213 (2013).
Betancur, C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380, 42–77 (2011).
Toro, R. et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 26, 363–372 (2010).
Phelan, K. & McDermid, H. E. The 22q13.3 deletion syndrome (Phelan-McDermid syndrome). Mol. Syndromol. 2, 186–201 (2012).
Phelan, M. C. et al. 22Q13 deletion syndrome. Am. J. Med. Genet. 101, 91–99 (2001).
Bonaglia, M. C. et al. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am. J. Hum. Genet. 69, 261–268 (2001). This study is the first to describe a possible association between PMS and disruption of the SHANK3 gene.
Bonaglia, M. C. et al. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with phelan/mcdermid syndrome. PLoS Genet. 7, e1002173 (2011).
Wilson, H. L. et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J. Med. Genet. 40, 575–584 (2003).
Wilson, H. L. et al. Interstitial 22q13 deletions: genes other than SHANK3 have major effects on cognitive and language development. Eur. J. Hum. Genet. 16, 1301–1310 (2008).
Moessner, R. et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 81, 1289–1297 (2007).
Durand, C. M. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27 (2007).
Berkel, S. et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 42, 489–491 (2010).
Okabe, S. Molecular anatomy of the postsynaptic density. Mol. Cell. Neurosci. 34, 503–518 (2007).
Emes, R. D. & Grant, S. G. N. Evolution of synapse complexity and diversity. Annu. Rev. Neurosci. 35, 111–131 (2012).
Boeckers, T. M. The postsynaptic density. Cell Tissue Res. 326, 409–422 (2006).
Montgomery, J. M., Zamorano, P. L. & Garner, C. C. MAGUKs in synapse assembly and function: an emerging view. Cell. Mol. Life Sci. 61, 911–929 (2004).
Sheng, M. & Kim, E. The Shank family of scaffold proteins. J. Cell Sci. 113, 1851–1856 (2000).
Tao-Cheng, J. H., Dosemeci, A., Gallant, P. E., Smith, C. & Reese, T. Activity induced changes in the distribution of Shanks at hippocampal synapses. Neuroscience 168, 11–17 (2010).
Tao-Cheng, J. H., Yang, Y., Reese, T. S. & Dosemeci, A. Differential distribution of shank and GKAP at the postsynaptic density. PLoS ONE 10, e0118750 (2015).
Wang, X. et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 20, 3093–3108 (2011).
Bozdagi, O. et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol. Autism 1, 15 (2010). This study describes the first Shank3 -mutant mice (exon 4 and exon 9 deletion). These mice show some ASD-relevant phenotypes.
Yang, M. et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J. Neurosci. 32, 6525–6541 (2012).
Kouser, M. et al. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J. Neurosci. 33, 18448–18468 (2013).
Zhou, Y. et al. Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron 89, 147–162 (2016).
Hung, A. Y. et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 28, 1697–1708 (2008). This study describes the first Shank1 -mutant mice (exon 14 and exon 15 deletion; PDZ domain deletion).
Won, H. et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486, 261–265 (2012). This study describes one of the first Shank2 -mutant mice (exon 6 and exon 7 deletion). These mice show autism-related behavioural phenotypes and NMDAR dysfunction.
Roussignol, G. et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 25, 3560–3570 (2005).
Sala, C. et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115–130 (2001).
Peixoto, R., Wang, W., Croney, D., Kozorovitskiy, Y. & Sabatini, B. Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B−/− mice. Nat. Neurosci. 19, 716–724 (2016).
Yi, F. et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science 352, aaf2669 (2016).
Shcheglovitov, A. et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 503, 267–271 (2013).
Gogolla, N., Takesian, A. E., Feng, G., Fagiolini, M. & Hensch, T. K. Sensory integration in mouse insular cortex reflects GABA circuit maturation. Neuron 83, 894–905 (2014).
Filice, F., Vörckel, K. J., Sungur, A. Ö., Wöhr, M. & Schwaller, B. Reduction in parvalbumin expression not loss of the parvalbumin-expressing GABA interneuron subpopulation in genetic parvalbumin and shank mouse models of autism. Mol. Brain 9, 10 (2016).
Lu, C. et al. Micro-electrode array recordings reveal reductions in both excitation and inhibition in cultured cortical neuron networks lacking Shank3. Mol. Psychiatry 21, 159–168 (2015).
Rubenstein, J. L. R. & Merzenich, M. M. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2, 255–267 (2003).
Bourgeron, T. A synaptic trek to autism. Curr. Opin. Neurobiol. 19, 231–234 (2009).
Bernhardt, B. C. & Singer, T. The neural basis of empathy. Annu. Rev. Neurosci. 35, 1–23 (2012).
Frith, C. D. The social brain? Phil. Trans. R. Soc. B 362, 671–678 (2007).
Lamm, C. & Singer, T. The role of anterior insular cortex in social emotions. Brain Struct. Funct. 214, 579–591 (2010).
Barak, B. & Feng, G. Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat. Neurosci. 19, 647–655 (2016).
Gauthier, J. et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl Acad. Sci. USA 107, 7863–7868 (2010).
Han, K. et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature 503, 72–77 (2013). This study shows that mice with SHANK3 overexpression exhibit synaptic dysfunction and manic-like phenotypes, thus reinforcing the idea that proper Shank3 gene dosage is crucial.
Silverman, J. L. et al. Sociability and motor functions in Shank1 mutant mice. Brain Res. 1380, 120–137 (2011).
Mao, W. et al. Shank1 regulates excitatory synaptic transmission in mouse hippocampal parvalbumin-expressing inhibitory interneurons. Eur. J. Neurosci. 41, 1025–1035 (2015).
Wöhr, M., Roullet, F. I., Hung, A. Y., Sheng, M. & Crawley, J. N. Communication impairments in mice lacking shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS ONE 6, e20631 (2011).
Lim, C.-S. et al. Enhancing inhibitory synaptic function reverses spatial memory deficits in Shank2 mutant mice. Neuropharmacology 112, 104–112 (2017).
Peter, S. et al. Dysfunctional cerebellar Purkinje cells contribute to autism-like behaviour in Shank2-deficient mice. Nat. Commun. 7, 12627 (2016).
Lord, C., Cook, E. H., Leventhal, B. L. & Amaral, D. G. Autism spectrum disorders. Neuron 28, 355–363 (2000).
Amaral, D., Geschwind, D. & Dawson, G. Autism Spectrum Disorders (Oxford Univ. Press, 2011).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106 (2014).
Guy, J., Gan, J., Selfridge, J., Cobb, S. & Bird, A. Reversal of neurological defects in a mouse model of Rett syndrome. Science 315, 1143–1147 (2007). This study shows for the first time a robust phenotypic reversal, both in immature and in mature adult animal models of Rett syndrome, by induced expression of MeCP2.
Garg, S. K. et al. Systemic delivery of MeCP2 rescues behavioral and cellular deficits in female mouse models of Rett syndrome. J. Neurosci. 33, 13612–13620 (2013).
Sztainberg, Y. et al. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 528, 123–126 (2015).
Clement, J. P. et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell 151, 709–723 (2012).
Bidinosti, M. et al. CLK2 inhibition ameliorates autistic features associated with SHANK3 deficiency. Science 351, 1199–1203 (2016).
Vicidomini, C. et al. Pharmacological enhancement of mGlu5 receptors rescues behavioral deficits in SHANK3 knock-out mice. Mol. Psychiatry http://dx.doi.org/10.1038/mp.2016.30 (2016).
Duffney, L. J. et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 11, 1400–1413 (2015).
Lee, E.-J. et al. Trans-synaptic zinc mobilization improves social interaction in two mouse models of autism through NMDAR activation. Nat. Commun. 6, 7168 (2015).
Jaramillo, T. C. et al. Altered striatal synaptic function and abnormal behaviour in Shank3 Exon4-9 deletion mouse model of autism. Autism Res. 9, 350–375 (2016).
Speed, H. E. et al. Autism-associated insertion mutation (InsG) of Shank3 Exon 21 causes impaired synaptic transmission and behavioral deficits. J. Neurosci. 35, 9648–9665 (2015).
Acknowledgements
The authors thank N. Chen for critical comments and editing the manuscript, N. Sousa (Minho University, Portugal) and all members of the Feng laboratory for support and helpful discussion. Research related to this work in the laboratory of G.F. is supported by the Poitras Center for Affective Disorders Research at the Massachusetts Institute of Technology (MIT), Stanley Center for Psychiatric Research at Broad Institute of MIT and Harvard, National Institute of Mental Health (MH097104), Nancy Lurie Marks Family Foundation, Simons Foundation Autism Research Initiative (SFARI grant 178130) and Simons Center for the Social Brain at MIT. P.M. is supported by Society in Science, The Branco Weiss Fellowship, administered by Eidgenössische Technische Hochschule (ETH) Zürich, and European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 89–2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Supplementary information
Supplementary information
Supplementary Figures and tables (PDF 12455 kb)
Glossary
- Intragenic promoters
-
Promoters located within the body of the residing gene, regulating its activity. Intragenic promoters are mainly active in tissue-specific gene expression.
- Alternative splicing
-
A process whereby different mRNAs can be produced from a single gene through the differential incorporation of exons into the mature transcript during splicing. Frequently, various mature proteins are generated from a single gene.
- Epigenetic mechanisms
-
A mechanism of a stable change in gene expression that does not involve changes in DNA sequence.
- Concordance
-
The occurrence of a trait in both of two related individuals, such as twins or siblings.
- Ring chromosomes
-
Structural aberrations of chromosomes, the long and short arms of which fuse together to form a ring. Ring chromosomes can have variable size and genetic content and are often associated with large terminal deletions.
- Interstitial deletions
-
The loss of one or more segments of DNA within a single chromosome (intra-chromosomal deletion), thereby joining genes that were previously far apart.
- Unbalanced translocations
-
Translocations are chromosomal abnormalities that are characterized by place exchange of parts of two or more different chromosomes. Balanced translocations have no net loss or gain of any chromosomal material, whereas unbalanced translocations have unequal exchange of chromosomal material, hence resulting in loss or gain of extra genes.
- Microdeletions
-
Small interstitial deletions of DNA (up to 5 Mb).
- Breakpoints
-
The specific sites of chromosomal breakage that are associated with a particular chromosomal rearrangement.
- Perineuronal nets
-
Aggregates of extracellular matrix molecules that embed cell bodies, axon initial segments and proximal dendrites of a subset of neurons in a mesh-like structure.
- Excitation/inhibition (E/I) ratio
-
The relationship between synaptic excitation and inhibition, two opposing forces in terms of neurotransmission.
- Multisensory integration
-
The neural processes that are involved in synthesizing information from cross-modal stimuli.
Rights and permissions
About this article
Cite this article
Monteiro, P., Feng, G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci 18, 147–157 (2017). https://doi.org/10.1038/nrn.2016.183
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn.2016.183
This article is cited by
-
Shank3 deficiency elicits autistic-like behaviors by activating p38α in hypothalamic AgRP neurons
Molecular Autism (2024)
-
Genetic architecture of childhood speech disorder: a review
Molecular Psychiatry (2024)
-
Neuron type-specific proteomics reveals distinct Shank3 proteoforms in iSPNs and dSPNs lead to striatal synaptopathy in Shank3B–/– mice
Molecular Psychiatry (2024)
-
Deficiency of FRMD5 results in neurodevelopmental dysfunction and autistic-like behavior in mice
Molecular Psychiatry (2024)
-
Joubert syndrome causing mutation in C2 domain of CC2D2A affects structural integrity of cilia and cellular signaling molecules
Experimental Brain Research (2024)