Abstract
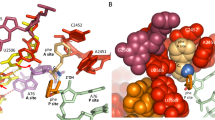
Protein synthesis is catalyzed in the peptidyl transferase center (PTC), located in the large (50S) subunit of the ribosome. No high-resolution structure of the intact ribosome has contained a complete active site including both A- and P-site tRNAs. In addition, although past structures of the 50S subunit have found no ordered proteins at the PTC, biochemical evidence suggests that specific proteins are capable of interacting with the 3′ ends of tRNA ligands. Here we present structures, at 3.6-Å and 3.5-Å resolution respectively, of the 70S ribosome in complex with A- and P-site tRNAs that mimic pre- and post-peptidyl-transfer states. These structures demonstrate that the PTC is very similar between the 50S subunit and the intact ribosome. They also reveal interactions between the ribosomal proteins L16 and L27 and the tRNA substrates, helping to elucidate the role of these proteins in peptidyl transfer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Nissen, P., Hansen, J., Ban, N., Moore, P.B. & Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Steitz, T.A. Structural insights into the functions of the large ribosomal subunit, a major antibiotic target. Keio J. Med. 57, 1–14 (2008).
Rodnina, M.V., Beringer, M. & Wintermeyer, W. Mechanism of peptide bond formation on the ribosome. Q. Rev. Biophys. 39, 203–225 (2006).
Korostelev, A., Trakhanov, S., Laurberg, M. & Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 (2006).
Bashan, A. et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11, 91–102 (2003).
Wower, I.K., Wower, J. & Zimmermann, R.A. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 273, 19847–19852 (1998).
Wower, J., Hixson, S.S. & Zimmermann, R.A. Labeling the peptidyl transferase center of the Escherichia coli ribosome with photoreactive tRNAPhe derivatives containing azidoadenosine at the 3′ end of the acceptor arm: a model of the tRNA-ribosome complex. Proc. Natl. Acad. Sci. USA 86, 5232–5236 (1989).
Maguire, B.A., Beniaminov, A.D., Ramu, H., Mankin, A.S. & Zimmermann, R.A. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol. Cell 20, 427–435 (2005).
Moore, V.G., Atchison, R.E., Thomas, G., Moran, M. & Noller, H.F. Identification of a ribosomal protein essential for peptidyl transferase activity. Proc. Natl. Acad. Sci. USA 72, 844–848 (1975).
Kazemie, M. Binding of aminoacyl-tRNA to reconstituted subparticles of Escherichia coli large ribosomal subunits. Eur. J. Biochem. 67, 373–378 (1976).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Kim, D.F. & Green, R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4, 859–864 (1999).
Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B. & Steitz, T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA 98, 4899–4903 (2001).
Schmeing, T.M., Huang, K.S., Strobel, S.A. & Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005).
Weinger, J.S., Parnell, K.M., Dorner, S., Green, R. & Strobel, S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 11, 1101–1106 (2004).
Dorner, S., Panuschka, C., Schmid, W. & Barta, A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 31, 6536–6542 (2003).
Wohlgemuth, I., Beringer, M. & Rodnina, M.V. Rapid peptide bond formation on isolated 50S ribosomal subunits. EMBO Rep. 7, 699–703 (2006).
Fahlman, R.P. & Uhlenbeck, O.C. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43, 7575–7583 (2004).
Fahlman, R.P., Dale, T. & Uhlenbeck, O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16, 799–805 (2004).
Semenkov, Y.P., Rodnina, M.V. & Wintermeyer, W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 7, 1027–1031 (2000).
Lill, R., Robertson, J.M. & Wintermeyer, W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 25, 3245–3255 (1986).
Schilling-Bartetzko, S., Franceschi, F., Sternbach, H. & Nierhaus, K.H. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J. Biol. Chem. 267, 4693–4702 (1992).
Zavialov, A.V., Mora, L., Buckingham, R.H. & Ehrenberg, M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 (2002).
Caskey, C.T., Beaudet, A.L., Scolnick, E.M. & Rosman, M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl. Acad. Sci. USA 68, 3163–3167 (1971).
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008).
Weixlbaumer, A. et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008).
Nierhaus, K.H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry 29, 4997–5008 (1990).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.S. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Maguire, B.A., Manuilov, A.V. & Zimmermann, R.A. Differential effects of replacing Escherichia coli ribosomal protein L27 with its homologue from Aquifex aeolicus. J. Bacteriol. 183, 6565–6572 (2001).
Chen, R., Mende, L. & Arfsten, U. The primary structure of protein L27 from the peptidyl-tRNA binding side of Escherichia coli ribosomes. FEBS Lett. 59, 96–99 (1975).
Odintsova, T.I. et al. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J. Protein Chem. 22, 249–258 (2003).
Strader, M.B. et al. Characterization of the 70S ribosome from Rhodopseudomonas palustris using an integrated “top-down” and “bottom-up” mass spectrometric approach. J. Proteome Res. 3, 965–978 (2004).
Suh, M.J., Hamburg, D.M., Gregory, S.T., Dahlberg, A.E. & Limbach, P.A. Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 5, 4818–4831 (2005).
Trobro, S. & Aqvist, J. Role of ribosomal protein L27 in peptidyl transfer. Biochemistry 47, 4898–4906 (2008).
Hampl, H., Schulze, H. & Nierhause, K.H. Ribosomal components from Escherichia coli 50S subunits involved in the reconstitution of peptidyl transferase activity. J. Biol. Chem. 258, 12810–12815 (1981).
Noller, H.F. et al. Structure of the ribosome at 5.5 A resolution and its interactions with functional ligands. Cold Spring Harb. Symp. Quant. Biol. 66, 57–66 (2001).
Nishimura, M. et al. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J. Mol. Biol. 344, 1369–1383 (2004).
Belova, L., Tenson, T., Xiong, L., McNicholas, P.M. & Mankin, A.S. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl. Acad. Sci. USA 98, 3726–3731 (2001).
Fraser, T.H. & Rich, A. Synthesis and aminoacylation of 3′-amino-3′-deoxy transfer RNA and its activity in ribosomal protein synthesis. Proc. Natl. Acad. Sci. USA 70, 2671–2675 (1973).
Robins, M.J., Miles, R.W., Samano, M.C. & Kaspar, R.L. Syntheses of puromycin from adenosine and 7-deazapuromycin from tubercidin, and biological comparisons of the 7-aza/deaza pair. J. Org. Chem. 66, 8204–8210 (2001).
Ludwig, J. A new route to nucleoside 5′-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung. 16, 131–133 (1981).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. D. Biol. Crystallogr. 54, 905–921 (1998).
Schmeing, T.M., Huang, K.S., Kitchen, D.E., Strobel, S.A. & Steitz, T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448 (2005).
Acknowledgements
We thank M. Schmeing for guidance with refinement and interpretation of the data, E. Stephens for mass spectrometry work, J.C. Cochrane for critical review of the manuscript, and M. Fuchs and C. Schulze-Briese for their advice and help with data collection at the Swiss Light Source. This work was supported by the Medical Research Council UK, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. R.M.V. is the recipient of a Gates-Cambridge scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
V.R. is on the Scientific Advisory Board of Rib-X Pharmaceuticals, a company involved in developing new antibiotics that target the ribosome.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 3253 kb)
Supplementary Video 1
Accommodation of the A-site tRNA. This movie models the conformational changes that occur within the 23S RNA upon A-site tRNA binding. The movie was made using coordinates from Selmer et al.1, which did not contain an A-site substrate within the PTC, and the pre-peptidyl transfer structure reported here, containing an ordered A-site tRNA within the PTC. 23S RNA is represented in cyan, A- and P-site tRNAs are modeled in green and red, respectively, and the A- and P-site amino acids are shown in yellow. (MOV 1224 kb)
Rights and permissions
About this article
Cite this article
Voorhees, R., Weixlbaumer, A., Loakes, D. et al. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16, 528–533 (2009). https://doi.org/10.1038/nsmb.1577
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1577
This article is cited by
-
Post-translational backbone-acyl shift yields natural product-like peptides bearing hydroxyhydrocarbon units
Nature Chemistry (2022)
-
Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling
Nature Communications (2021)
-
Evolution of ribosomal protein network architectures
Scientific Reports (2021)
-
Peptidyl transferase center decompaction and structural constraints during early protein elongation on the ribosome
Scientific Reports (2021)
-
trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo
Nature Communications (2021)