Abstract
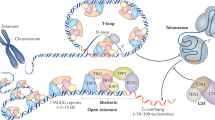
To maintain linear DNA genomes, organisms have evolved numerous means of solving problems associated with DNA ends (telomeres), including telomere-associated retrotransposons, palindromes, hairpins, covalently bound proteins and the addition of arrays of simple DNA repeats. Telomeric arrays can be maintained through various mechanisms such as telomerase activity or recombination. The recombination-dependent maintenance pathways may include telomeric loops (t-loops) and telomeric circles (t-circles). The potential involvement of t-circles in telomere maintenance was first proposed for linear mitochondrial genomes. The occurrence of t-circles in a wide range of organisms, spanning yeasts, plants and animals, suggests the involvement of t-circles in many phenomena including the alternative-lengthening of telomeres (ALT) pathway and telomere rapid deletion (TRD). In this Perspective, we summarize these findings and discuss how t-circles may be related to t-loops and how t-circles may have initiated the evolution of telomeres.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gilson, E. & Géli, V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 (2007).
McEachern, M.J., Krauskopf, A. & Blackburn, E.H. Telomeres and their control. Annu. Rev. Genet. 34, 331–358 (2000).
Verdun, R.E. & Karlseder, J. Replication and protection of telomeres. Nature 447, 924–931 (2007).
Nosek, J., Kosa, P. & Tomaska, L. On the origin of telomeres: a glimpse at the pre-telomerase world. Bioessays 28, 182–190 (2006).
Pardue, M.L. & DeBaryshe, P.G. Drosophila telomeres: a variation on the telomerase theme. in Origin and Evolution of Telomeres (eds. Nosek, J. & Tomaska, L.) 27–44 (Landes Bioscience, Austin, Texas, 2008).
Pich, U., Fuchs, J. & Schubert, I. How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res. 4, 207–213 (1996).
Nosek, J. & Tomaska, L. Mitochondrial telomeres: an evolutionary paradigm for the emergence of telomeric structures and their replication strategies. in Origin and Evolution of Telomeres (eds. Nosek, J. & Tomaska, L.) 163–171 (Landes Bioscience, Austin, Texas, 2008).
Tomaska, L., McEachern, M.J. & Nosek, J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 567, 142–146 (2004).
Blackburn, E.H. Telomere states and cell fates. Nature 408, 53–56 (2000).
de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005).
Smogorzewska, A. & de Lange, T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 (2004).
Cesare, A.J. & Reddel, R.R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129, 99–108 (2008).
Cesare, A.J. & Reddel, R.R. Alternative lengthening of telomeres in mammalian cells. in Origin and Evolution of Telomeres (eds. Nosek, J. & Tomaska, L.) 45–57 (Landes Bioscience, Austin, Texas, 2008).
Lundblad, V. Telomere maintenance without telomerase. Oncogene 21, 522–531 (2002).
Lundblad, V. & Blackburn, E.H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73, 347–360 (1993).
Teng, S.C. & Zakian, V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8083–8093 (1999).
Kovac, L., Lazowska, J. & Slonimski, P.P. A yeast with linear molecules of mitochondrial DNA. Mol. Gen. Genet. 197, 420–424 (1984).
Nosek, J., Dinouel, N., Kovac, L. & Fukuhara, H. Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol. Gen. Genet. 247, 61–72 (1995).
Tomaska, L., Nosek, J. & Fukuhara, H. Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J. Biol. Chem. 272, 3049–3056 (1997).
Nosek, J., Tomaska, L., Pagacova, B. & Fukuhara, H. Mitochondrial telomere-binding protein from Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA-binding protein. J. Biol. Chem. 274, 8850–8857 (1999).
Tomaska, L., Makhov, A.M., Nosek, J., Kucejova, B. & Griffith, J.D. Electron microscopic analysis supports a dual role for the mitochondrial telomere-binding protein of Candida parapsilosis. J. Mol. Biol. 305, 61–69 (2001).
Tomaska, L., Nosek, J. & Kucejova, B. Mitochondrial single-stranded DNA-binding proteins: in search for new functions. Biol. Chem. 382, 179–186 (2001).
Tomaska, L., Nosek, J., Makhov, A.M., Pastorakova, A. & Griffith, J.D. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28, 4479–4487 (2000).
Nosek, J., Rycovska, A., Makhov, A.M., Griffith, J.D. & Tomaska, L. Amplification of telomeric arrays via rolling-circle mechanism. J. Biol. Chem. 280, 10840–10845 (2005).
Kosa, P., Valach, M., Tomaska, L., Wolfe, K.H. & Nosek, J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 34, 2472–2481 (2006).
Rycovska, A., Valach, M., Tomaska, L., Bolotin-Fukuhara, M. & Nosek, J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology 150, 1571–1580 (2004).
Cohen, S., Regev, A. & Lavi, S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene 14, 977–985 (1997).
Ogino, H. et al. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 248, 223–227 (1998).
Regev, A., Cohen, S., Cohen, E., Bar-Am, I. & Lavi, S. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene 17, 3455–3461 (1998).
Tokutake, Y. et al. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun. 247, 765–772 (1998).
Cesare, A.J., Groff-Vindman, C., Compton, S.A., McEachern, M.J. & Griffith, J.D. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell. Biol. 28, 20–29 (2008).
Groff-Vindman, C., Cesare, A.J., Natarajan, S., Griffith, J.D. & McEachern, M.J. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 25, 4406–4412 (2005).
Raices, M. et al. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132, 745–757 (2008).
Zellinger, B., Akimcheva, S., Puizina, J., Schirato, M. & Riha, K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27, 163–169 (2007).
Cohen, S. & Méchali, M. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 3, 1168–1174 (2002).
Cesare, A.J. & Griffith, J.D. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24, 9948–9957 (2004).
Nabetani, A. & Ishikawa, F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 29, 703–713 (2009).
Wang, R.C., Smogorzewska, A. & de Lange, T. Homologous recombination generates t-loop-sized deletions at human telomeres. Cell 119, 355–368 (2004).
Wang, Y., Ghosh, G. & Hendrickson, E.A. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA 106, 12430–12435 (2009).
Pickett, H.A., Cesare, A.J., Johnston, R.L., Neumann, A.A. & Reddel, R.R. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28, 799–809 (2009).
Liu, L. et al. Telomere lengthening early in development. Nat. Cell Biol. 9, 1436–1441 (2007).
Topcu, Z., Nickles, K., Davis, C. & McEachern, M.J. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl. Acad. Sci. USA 102, 3348–3353 (2005).
Natarajan, S., Groff-Vindman, C. & McEachern, M.J. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell 2, 1115–1127 (2003).
Natarajan, S. & McEachern, M.J. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22, 4512–4521 (2002).
Tomaska, L. & Nosek, J. Telomere heterogeneity: taking advantage of stochastic events. FEBS Lett. 583, 1067–1071 (2009).
Lustig, A.J. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4, 916–923 (2003).
Cerone, M.A., Londono-Vallejo, J.A. & Bacchetti, S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum. Mol. Genet. 10, 1945–1952 (2001).
Deng, Z., Dheekollu, J., Broccoli, D., Dutta, A. & Lieberman, P.M. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 17, 1989–1995 (2007).
Li, B., Jog, S.P., Reddy, S. & Comai, L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2ΔB-mediated telomere shortening. Mol. Cell. Biol. 28, 1892–1904 (2008).
Compton, S.A., Choi, J.H., Cesare, A.J., Ozgur, S. & Griffith, J.D. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 67, 1513–1519 (2007).
Zhong, Z.H. et al. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 282, 29314–29322 (2007).
Cerone, M.A., Autexier, C., Londono-Vallejo, J.A. & Bacchetti, S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene 24, 7893–7901 (2005).
Jiang, W.Q. et al. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 25, 2708–2721 (2005).
Muntoni, A. & Reddel, R.R. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14, R191–R196 (2005).
Compton, S.A., Tolun, G., Kamath-Loeb, A.S., Loeb, L.A. & Griffith, J.D. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 283, 24478–24483 (2008).
Poulet, A. et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 28, 641–651 (2009).
Fouché, N. et al. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 281, 37486–37495 (2006).
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
de Lange, T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5, 323–329 (2004).
Blanc, H. & Dujon, B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl. Acad. Sci. USA 77, 3942–3946 (1980).
MacAlpine, D.M., Kolesar, J., Okamoto, K., Butow, R.A. & Perlman, P.S. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 20, 1807–1817 (2001).
Bibillo, A. & Eickbush, T.H. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 279, 14945–14953 (2004).
Chen, B. & Lambowitz, A.M. De novo and DNA primer-mediated initiation of cDNA synthesis by the mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J. Mol. Biol. 271, 311–332 (1997).
Kennell, J.C., Wang, H. & Lambowitz, A.M. The Mauriceville plasmid of Neurospora spp. uses novel mechanisms for initiating reverse transcription in vivo. Mol. Cell. Biol. 14, 3094–3107 (1994).
Azzalin, C.M., Reichenbach, P., Khoriauli, L., Giulotto, E. & Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007).
Schoeftner, S. & Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 (2008).
Nosek, J. et al. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol. Genet. Genomics 272, 173–180 (2004).
Tomaska, L., Makhov, A.M., Griffith, J.D. & Nosek, J. t-loops in yeast mitochondria. Mitochondrion 1, 455–459 (2002).
Acknowledgements
We wish to thank L. Kovac (Comenius University) for inspiration and continuous support and members of our laboratories for discussions. We also thank two anonymous reviewers for valuable comments and suggestions. Our work related to telomere biology is supported by grants from a Fogarty International Research Collaboration Award (2-R03-TW005654-04A1), the Howard Hughes Medical Institute (55005622), the Slovak grant agencies APVT (20-001604 and 0024-07) and VEGA (1/0132/09 and 1/0219/08), and grants to J.D.G. (US National Institutes of Health grants GM31819 and ES13773 and awards from the Ellison and Glenn foundations).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Tomaska, L., Nosek, J., Kramara, J. et al. Telomeric circles: universal players in telomere maintenance?. Nat Struct Mol Biol 16, 1010–1015 (2009). https://doi.org/10.1038/nsmb.1660
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1660
This article is cited by
-
Extrachromosomal circular DNA: a new potential role in cancer progression
Journal of Translational Medicine (2021)
-
Telomere damage induces internal loops that generate telomeric circles
Nature Communications (2020)
-
Chromosomal integration of HHV-6A during non-productive viral infection
Scientific Reports (2017)
-
Telomere recombination pathways: tales of several unhappy marriages
Current Genetics (2017)
-
Alternative lengthening of telomeres: models, mechanisms and implications
Nature Reviews Genetics (2010)