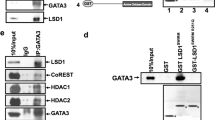
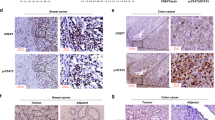
Abstract
ID4 (inhibitor of DNA binding 4) is a member of a family of proteins that function as dominant-negative regulators of basic helix-loop-helix transcription factors. Growing evidence links ID proteins to cell proliferation, differentiation and tumorigenesis. Here we identify ID4 as a transcriptional target of gain-of-function p53 mutants R175H, R273H and R280K. Depletion of mutant p53 protein severely impairs ID4 expression in proliferating tumor cells. The protein complex mutant p53–E2F1 assembles on specific regions of the ID4 promoter and positively controls ID4 expression. The ID4 protein binds to and stabilizes mRNAs encoding pro-angiogenic factors IL8 and GRO-α. This results in the increase of the angiogenic potential of cancer cells expressing mutant p53. These findings highlight the transcriptional axis mutant p53, E2F1 and ID4 as a still undefined molecular mechanism contributing to tumor neo-angiogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Norton, J.D. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 113, 3897–3905 (2000).
Yokota, Y. & Mori, S. Role of Id family proteins in growth control. J. Cell. Physiol. 190, 21–28 (2002).
Ruzinova, M.B. & Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13, 410–418 (2003).
Lasorella, A., Uo, T. & Iavarone, A. Id proteins at the cross-road of development and cancer. Oncogene 20, 8326–8333 (2001).
Fong, S., Debs, R.J. & Desprez, P.Y. Id genes and proteins as promising targets in cancer therapy. Trends Mol. Med. 10, 387–392 (2004).
Perk, J., Iavarone, A. & Benezra, R. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5, 603–614 (2005).
Beger, C. et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library–based inverse genomics approach. Proc. Natl. Acad. Sci. USA 98, 130–135 (2001).
Welcsh, P.L. et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc. Natl. Acad. Sci. USA 99, 7560–7565 (2002).
Shan, L., Yu, M., Qiu, C. & Snyderwine, E.G. Id4 regulates mammary epithelial cell growth and differentiation and is overexpressed in rat mammary gland carcinomas. Am. J. Pathol. 163, 2495–2502 (2003).
de Candia, P., Akram, M., Benezra, R. & Brogi, E. Id4 messenger RNA and estrogen receptor expression: inverse correlation in human normal breast epithelium and carcinoma. Hum. Pathol. 37, 1032–1041 (2006).
Sigal, A. & Rotter, V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 60, 6788–6793 (2000).
Cadwell, C. & Zambetti, G.P. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene 277, 15–30 (2001).
Gualberto, A., Aldape, K., Kozakiewicz, K. & Tlsty, T.D. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc. Natl. Acad. Sci. USA 95, 5166–5171 (1998).
Murphy, K.L., Dennis, A.P. & Rosen, J.M. A gain of function p53 mutant promotes both genomic instability and cell survival in a novel p53-null mammary epithelial cell model. FASEB J. 14, 2291–2302 (2000).
Lotem, J. & Sachs, L.A. Mutant p53 antagonizes the deregulated c-myc-mediated enhancement of apoptosis and decrease in leukemogenicity. Proc. Natl. Acad. Sci. USA 92, 9672–9676 (1995).
Li, R. et al. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene 16, 3269–3277 (1998).
Blandino, G., Levine, A.J. & Oren, M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18, 477–485 (1999).
Matas, D. et al. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20, 4163–4172 (2001).
Bossi, G. et al. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 25, 304–309 (2006).
Strano, S. et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 275, 29503–29512 (2000).
Gaiddon, C., Lokshin, M., Ahn, J., Zhang, T. & Prives, C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21, 1874–1887 (2001).
Lin, J., Teresky, A.K. & Levine, A.J. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene 10, 2387–2390 (1995).
Kim, E. & Deppert, W. Transcriptional activities of mutant p53: when mutations are more than a loss. J. Cell. Biochem. 93, 878–886 (2004).
Di Agostino, S. et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10, 191–202 (2006).
Strano, S. et al. Mutant p53 proteins: between loss and gain of function. Head Neck 29, 488–496 (2007).
Weisz, L., Oren, M. & Rotter, V. Transcription regulation by mutant p53. Oncogene 26, 2202–2211 (2007).
Rhodes, D.R. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 (2004).
Neve, R.M. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006).
Liu, N., Lucibello, F.C., Engeland, K. & Muller, R. A new model of cell cycle-regulated transcription: repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene 16, 2957–2963 (1998).
Fogal, V., Hsieh, K., Royer, C., Zhong, S. & Lu, X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J. 24, 2768–2782 (2005).
Qin, G. et al. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proc. Natl. Acad. Sci. USA 103, 11015–11020 (2006).
Caunt, M. et al. Growth-regulated oncogene is pivotal in thrombin-induced angiogenesis. Cancer Res. 66, 4125–4132 (2006).
Waugh, D.J. & Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 14, 6735–6741 (2008).
Anderson, P. Post-transcriptional control of cytokine production. Nat. Immunol. 9, 353–359 (2008).
Datta, S. et al. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J. Immunol. 180, 2545–2552 (2008).
Vasudevan, S. & Steitz, J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128, 1105–1118 (2007).
Espel, E. The role of the AU-rich elements of mRNAs in controlling translation. Semin. Cell Dev. Biol. 16, 59–67 (2005).
Barreau, C., Paillard, L. & Osborne, H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 (2005).
Zalcenstein, A. et al. Repression of the MSP/MST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene 25, 359–369 (2006).
Zalcenstein, A. et al. Mutant p53 gain of function: repression of CD95 (Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene 22, 5667–5676 (2003).
Weisz, L. et al. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 64, 8318–8327 (2004).
Gualberto, A. & Baldwin, A.S. Jr. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J. Biol. Chem. 270, 19680–19683 (1995).
Chicas, A., Molina, P. & Bargonetti, J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem. Biophys. Res. Commun. 279, 383–390 (2000).
Sampath, J. et al. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J. Biol. Chem. 276, 39359–39367 (2001).
Lyden, D. et al. ID1 and ID3 are required for neurogenesis, angiogenesis and vascularization of tumor xenografts. Nature 401, 670–677 (1999).
Linderholm, B.K. et al. The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res. 61, 2256–2260 (2001).
Fontemaggi, G. et al. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J. Biol. Chem. 277, 43359–43368 (2002).
Pagliuca, A., Cannada-Bartoli, P. & Lania, L. A role for Sp and helix-loop-helix transcription factors in the regulation of the human Id4 gene promoter activity. J. Biol. Chem. 273, 7668–7674 (1998).
Samanta, J. & Kessler, J.A. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131, 4131–4142 (2004).
Brummelkamp, T.R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Fontemaggi, G. et al. The transcriptional repressor ZEB regulates p73 expression at the crossroad between proliferation and differentiation. Mol. Cell. Biol. 21, 8461–8470 (2001).
Sanvito, F. et al. The β4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 144, 823–837 (1999).
Acknowledgements
We are grateful to J.A. Kessler (Northwestern University's Feinberg School of Medicine) and L. Lania (University of Naples Federico II) for providing plasmids. We also thank G. Starace (Italian National Research Council (CNR)) for cell sorting. This work was supported by European Community (EC) FP6 “Active p53” and “Mutant p53” consortia. This publication reflects the authors' views and not necessarily those of the EC. The EC is not liable for any use that may be made of the information contained herein. Support by Associazione Italiana per la Ricerca sul Cancro (AIRC)-Rome Oncogenic Centre (ROC) to the oncogenomic platform, AIRC to G.B., S.S., F.F. and D.D.B., Lega Italiana Tumori to S.S., Ministero della Sanità and Alleanza contro il cancro is greatly appreciated. S.D.O. holds a fellowship from Italian Association for Cancer Research (FIRC).
Author information
Authors and Affiliations
Contributions
G.F. and S.D.O. performed microarrays, transcriptional and mRNA stability studies, chromatin and ribonucleoprotein immunoprecipitations; E.D. and T.S. performed bioinformatic analysis of microarray data; F.F. performed studies on polysomal cell fractions; M.M. and E.M. performed stainings on breast cancer specimens; I.T. and P.M. performed statistical analysis of breast cancer immunohistochemical data; D.D.B. and D.T. performed angiogenic assays. G.B., G.F. and S.S. supervised the project.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–3 and Supplementary Methods (PDF 706 kb)
Rights and permissions
About this article
Cite this article
Fontemaggi, G., Dell'Orso, S., Trisciuoglio, D. et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol 16, 1086–1093 (2009). https://doi.org/10.1038/nsmb.1669
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1669
This article is cited by
-
ID4-dependent secretion of VEGFA enhances the invasion capability of breast cancer cells and activates YAP/TAZ via integrin β3-VEGFR2 interaction
Cell Death & Disease (2024)
-
Mutant p53 in cancer: from molecular mechanism to therapeutic modulation
Cell Death & Disease (2022)
-
Should mutant TP53 be targeted for cancer therapy?
Cell Death & Differentiation (2022)
-
Id proteins: emerging roles in CNS disease and targets for modifying neural stemcell behavior
Cell and Tissue Research (2022)
-
CDC42 as an epigenetic regulator of ID4 in triple-negative breast tumors
Breast Cancer (2022)