Abstract
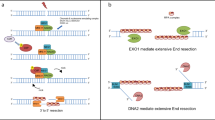
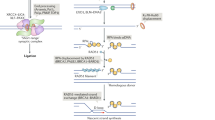
DNA double-strand breaks are repaired by different mechanisms, including homologous recombination and nonhomologous end-joining. DNA-end resection, the first step in recombination, is a key step that contributes to the choice of DSB repair. Resection, an evolutionarily conserved process that generates single-stranded DNA, is linked to checkpoint activation and is critical for survival. Failure to regulate and execute this process results in defective recombination and can contribute to human disease. Here I review recent findings on the mechanisms of resection in eukaryotes, from yeast to vertebrates, provide insights into the regulatory strategies that control it, and highlight the consequences of both its impairment and its deregulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Aguilera, A. & Gómez-González, B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217 (2008).
Lieber, M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 (2008).
Krogh, B.O. & Symington, L.S. Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 (2004).
McVey, M. & Lee, S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538 (2008).
Dynan, W.S. & Yoo, S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26, 1551–1559 (1998).
Huertas, P., Cortes-Ledesma, F., Sartori, A.A., Aguilera, A. & Jackson, S.P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008).
Lee, K., Zhang, Y. & Lee, S. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454, 543–546 (2008).
Huertas, P. & Jackson, S.P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009).
Yun, M.H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Harrison, J.C. & Haber, J.E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 (2006).
Jazayeri, A. et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45 (2006).
Amundsen, S.K. & Smith, G.R. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112, 741–744 (2003).
Gravel, S., Chapman, J.R., Magill, C. & Jackson, S.P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 (2008).
Khakhar, R.R., Cobb, J., Bjergbaek, L., Hickson, I. & Gasser, S.M. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 13, 493–501 (2003).
Limbo, O. et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 (2007).
Mimitou, E.P. & Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Penkner, A. et al. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 26, 5071–5082 (2007).
Sartori, A.A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Tran, P.T., Erdeniz, N., Symington, L.S. & Liskay, R.M. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst.) 3, 1549–1559 (2004).
Uanschou, C. et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26, 5061–5070 (2007).
Zhu, Z., Chung, W.H., Shim, E., Lee, S. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Williams, R.S., Williams, J.S. & Tainer, J.A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 (2007).
Paull, T.T. & Gellert, M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 (1998).
Bressan, D.A., Olivares, H.A., Nelms, B.E. & Petrini, J.H. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150, 591–600 (1998).
Lavin, M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26, 7749–7758 (2007).
Buis, J. et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96 (2008).
Trujillo, K.M., Yuan, S.S., Lee, E.Y. & Sung, P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273, 21447–21450 (1998).
Falck, J., Coates, J.A. & Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 (2005).
Nakada, D., Matsumoto, K. & Sugimoto, K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17, 1957–1962 (2003).
Usui, T., Petrini, J.H. & Morales, M. rad50S alleles of the Mre11 complex: questions answered and questions raised. Exp. Cell Res. 312, 2694–2699 (2006).
McKee, A.H. & Kleckner, N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146, 797–816 (1997).
Neale, M.J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein–linked DNA double-strand breaks. Nature 436, 1053–1057 (2005).
Prinz, S., Amon, A. & Klein, F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146, 781–795 (1997).
Clerici, M., Mantiero, D., Lucchini, G. & Longhese, M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280, 38631–38638 (2005).
Lengsfeld, B.M., Rattray, A.J., Bhaskara, V., Ghirlando, R. & Paull, T.T. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651 (2007).
Jazayeri, A., Balestrini, A., Garner, E., Haber, J.E. & Costanzo, V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 27, 1953–1962 (2008).
Maringele, L. & Lydall, D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18, 2663–2675 (2004).
Mantiero, D., Clerici, M., Lucchini, G. & Longhese, M.P. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8, 380–387 (2007).
Moreau, S., Morgan, E.A. & Symington, L.S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159, 1423–1433 (2001).
Bae, S.H. et al. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273, 26880–26890 (1998).
Horejsí, Z. et al. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23, 3122–3127 (2004).
Chen, L., Nievera, C.J., Lee, A.Y. & Wu, X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283, 7713–7720 (2008).
Yu, X., Fu, S., Lai, M., Baer, R. & Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006).
Nimonkar, A.V., Ozsoy, A.Z., Genschel, J., Modrich, P. & Kowalczykowski, S.C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA 105, 16906–16911 (2008).
Kim, J.H. et al. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 34, 1854–1864 (2006).
Duxin, J.P. et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 29, 4274–4282 (2009).
Liao, S., Toczylowski, T. & Yan, H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′→3′ strand-specific processing of DNA ends. Nucleic Acids Res. 36, 6091–6100 (2008).
Hopkins, B.B. & Paull, T.T. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135, 250–260 (2008).
Zheng, L. et al. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 37, 2645–2657 (2009).
Aylon, Y., Liefshitz, B. & Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 (2004).
Ira, G. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 (2004).
Zierhut, C. & Diffley, J.F. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27, 1875–1885 (2008).
Gu, B. & Chen, P.L. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle 8, 1409–1420 (2009).
Lazzaro, F. et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27, 1502–1512 (2008).
Grenon, M. et al. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24, 105–119 (2007).
Linding, R. et al. Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 (2007).
Akamatsu, Y. et al. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 28, 3639–3651 (2008).
Yu, X. & Chen, J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24, 9478–9486 (2004).
Lisby, M., Barlow, J.H., Burgess, R.C. & Rothstein, R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004).
Clerici, M., Mantiero, D., Guerini, I., Lucchini, G. & Longhese, M.P. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9, 810–818 (2008).
Barlow, J.H., Lisby, M. & Rothstein, R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30, 73–85 (2008).
Baroni, E., Viscardi, V., Cartagena-Lirola, H., Lucchini, G. & Longhese, M.P. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24, 4151–4165 (2004).
Warmerdam, D.O., Freire, R., Kanaar, R. & Smits, V.A. Cell cycle-dependent processing of DNA lesions controls localization of Rad9 to sites of genotoxic stress. Cell Cycle 8, 1765–1774 (2009).
Shimada, K., Pasero, P. & Gasser, S.M. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16, 3236–3252 (2002).
Morin, I. et al. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 27, 2400–2410 (2008).
Zhang, Y. et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14, 639–646 (2007).
Chen, L. et al. Effect of amino acid substitutions in the Rad50 ATP binding domain on DNA double strand break repair in yeast. J. Biol. Chem. 280, 2620–2627 (2005).
Stracker, T.H., Theunissen, J.W., Morales, M. & Petrini, J.H. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst.) 3, 845–854 (2004).
Chen, P.L. et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25, 3535–3542 (2005).
Waltes, R. et al. Human RAD50 deficiency in an Nijmegen Breakage Syndrome-like disorder. Am. J. Hum. Genet. 84, 605–616 (2009).
Chinnadurai, G. CtIP, a candidate tumor susceptibility gene is a team player with luminaries. Biochim. Biophys. Acta 1765, 67–73 (2006).
Wu, G. & Lee, W.H. CtIP, a multivalent adaptor connecting transcriptional regulation, checkpoint control and tumor suppression. Cell Cycle 5, 1592–1596 (2006).
Cheok, C.F. et al. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 33, 1456–1459 (2005).
Lord, C.J. & Ashworth, A. Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 8, 363–969 (2008).
Acknowledgements
I would like to apologize to all authors whose work could not be cited due to space limitations. I am grateful to all the members of S. Jackson's laboratory in Cambridge, UK, for helpful discussions and especially to S. Jackson, A. Kaidi, J. Harrigan, K. Miller and R. Belotserkovskaya for their helpful suggestions and comments on the manuscript. I would also like to thank BBSRC and Cancer Research UK for funding my work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huertas, P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol 17, 11–16 (2010). https://doi.org/10.1038/nsmb.1710
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1710
This article is cited by
-
Impact of NAD+ metabolism on ovarian aging
Immunity & Ageing (2023)
-
ABRAXAS1 orchestrates BRCA1 activities to counter genome destabilizing repair pathways—lessons from breast cancer patients
Cell Death & Disease (2023)
-
Strategies to improve homology-based repair outcomes following CRISPR-based gene editing in mosquitoes: lessons in how to keep any repair disruptions local
Virology Journal (2022)
-
Wee1 promotes cell proliferation and imatinib resistance in chronic myeloid leukemia via regulating DNA damage repair dependent on ATM-γH2AX-MDC1
Cell Communication and Signaling (2022)
-
Development of transgenic Daphnia magna for visualizing homology-directed repair of DNA
Scientific Reports (2022)