Abstract
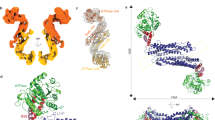
Mitochondria are dynamic organelles that undergo cycles of fission and fusion. The yeast dynamin-related protein Dnm1 has been localized to sites of mitochondrial division. Using cryo-EM, we have determined the three-dimensional (3D) structure of Dnm1 in a GTP-bound state. The 3D map showed that Dnm1 adopted a unique helical assembly when compared with dynamin, which is involved in vesicle scission during endocytosis. Upon GTP hydrolysis, Dnm1 constricted liposomes and subsequently dissociated from the lipid bilayer. The magnitude of Dnm1 constriction was substantially larger than the decrease in diameter previously reported for dynamin. We postulate that the larger conformational change is mediated by a flexible Dnm1 structure that has limited interaction with the underlying bilayer. Our structural studies support the idea that Dnm1 has a mechanochemical role during mitochondrial division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hoppins, S., Lackner, L. & Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 (2007).
Smirnova, E., Shurland, D.L., Ryazantsev, S.N. & van der Bliek, A.M. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358 (1998).
Bleazard, W. et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 (1999).
Knott, A.B., Perkins, G., Schwarzenbacher, R. & Bossy-Wetzel, E. Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 9, 505–518 (2008).
Frank, S. et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1, 515–525 (2001).
Arimura, S. & Tsutsumi, N. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99, 5727–5731 (2002).
Otsuga, D. et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349 (1998).
Smirnova, E., Griparic, L., Shurland, D.L. & van der Bliek, A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 (2001).
Mozdy, A.D., McCaffery, J.M. & Shaw, J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 (2000).
Griffin, E.E., Graumann, J. & Chan, D.C. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170, 237–248 (2005).
Tieu, Q., Okreglak, V., Naylor, K. & Nunnari, J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452 (2002).
James, D.I., Parone, P.A., Mattenberger, Y. & Martinou, J.C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 278, 36373–36379 (2003).
Heymann, J.A. & Hinshaw, J.E. Dynamins at a glance. J. Cell Sci. 122, 3427–3431 (2009).
Praefcke, G.J. & McMahon, H.T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 (2004).
Chappie, J.S., Acharya, S., Leonard, M., Schmid, S.L. & Dyda, F. G domain dimerization controls dynamin′s assembly-stimulated GTPase activity. Nature (2010).
Chappie, J.S. et al. An intramolecular signaling element that modulates dynamin function in vitro and in vivo. Mol. Biol. Cell 20, 3561–3571 (2009).
Muhlberg, A.B., Warnock, D.E. & Schmid, S.L. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 16, 6676–6683 (1997).
Song, B.D., Yarar, D. & Schmid, S.L. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol. Cell 15, 2243–2252 (2004).
Gao, S. et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010).
Ingerman, E. et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 (2005).
Low, H.H., Sachse, C., Amos, L.A. & Lowe, J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342–1352 (2009).
Okamoto, P.M., Tripet, B., Litowski, J., Hodges, R.S. & Vallee, R.B. Multiple distinct coiled-coils are involved in dynamin self-assembly. J. Biol. Chem. 274, 10277–10286 (1999).
Ramachandran, R. et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 26, 559–566 (2007).
Smirnova, E., Shurland, D.L., Newman-Smith, E.D., Pishvaee, B. & van der Bliek, A.M. A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem. 274, 14942–14947 (1999).
Klein, D.E., Lee, A., Frank, D.W., Marks, M.S. & Lemmon, M.A. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J. Biol. Chem. 273, 27725–27733 (1998).
Zheng, J. et al. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J. Mol. Biol. 255, 14–21 (1996).
Accola, M.A., Huang, B., Al Masri, A. & McNiven, M.A. The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277, 21829–21835 (2002).
Hinshaw, J.E. & Schmid, S.L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192 (1995).
Kim, Y.W. et al. Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol. 127, 1243–1255 (2001).
Carr, J.F. & Hinshaw, J.E. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J. Biol. Chem. 272, 28030–28035 (1997).
Kochs, G., Haener, M., Aebi, U. & Haller, O. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277, 14172–14176 (2002).
Yoon, Y., Pitts, K.R. & McNiven, M.A. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905 (2001).
Sweitzer, S.M. & Hinshaw, J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021–1029 (1998).
Takei, K., Slepnev, V.I., Haucke, V. & De Camilli, P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1, 33–39 (1999).
Vallis, Y., Wigge, P., Marks, B., Evans, P.R. & McMahon, H.T. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 9, 257–260 (1999).
Danino, D., Moon, K.H. & Hinshaw, J.E. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 147, 259–267 (2004).
Chen, Y.J., Zhang, P., Egelman, E.H. & Hinshaw, J.E. The stalk region of dynamin drives the constriction of dynamin tubes. Nat. Struct. Mol. Biol. 11, 574–575 (2004).
Zhang, P. & Hinshaw, J.E. Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 3, 922–926 (2001).
Mears, J.A., Ray, P. & Hinshaw, J.E. A corkscrew model for dynamin constriction. Structure 15, 1190–1202 (2007).
Lackner, L.L., Horner, J.S. & Nunnari, J. Mechanistic analysis of a dynamin effector. Science 325, 874–877 (2009).
Mears, J.A. & Hinshaw, J.E. Visualization of dynamins. Methods Cell Biol. 88, 237–256 (2008).
Egelman, E.H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000).
Niemann, H.H., Knetsch, M.L., Scherer, A., Manstein, D.J. & Kull, F.J. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 20, 5813–5821 (2001).
Doherty, G.J. & McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 (2009).
Ferguson, S.M. et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev. Cell 17, 811–822 (2009).
Roux, A. et al. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. USA 107, 4141–4146 (2010).
Bashkirov, P.V. et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135, 1276–1286 (2008).
Kozlovsky, Y. & Kozlov, M.M. Membrane fission: model for intermediate structures. Biophys. J. 85, 85–96 (2003).
Pucadyil, T.J. & Schmid, S.L. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 135, 1263–1275 (2008).
Roux, A., Uyhazi, K., Frost, A. & De Camilli, P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441, 528–531 (2006).
Zhang, Y. & Chan, D.C. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. USA 104, 18526–18530 (2007).
Johnston, C.A., Kimple, A.J., Giguere, P.M. & Siderovski, D.P. Structure of the parathyroid hormone receptor C terminus bound to the G-protein dimer Gbeta1gamma2. Structure 16, 1086–1094 (2008).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Kremer, J.R., Mastronarde, D.N. & McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank D. Winkler and A. Steven for technical assistance in acquiring tomograms and P. Flicker, J. Hanover, W. Prinz, J. Evans and J. Heymann for critical discussions and reading of the manuscript. This work was supported by the Intramural Research Program of the US National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (J.E.H.) and the Nancy Nossal Fellowship of the NIH, NIDDK (J.A.M.). J.N. is supported by NIH grant R01GM062942, and L.L.L. is supported by NIH postdoctoral fellowship 1F32GM078749.
Author information
Authors and Affiliations
Contributions
J.A.M. prepared, imaged and processed the cryo-EM data. L.L.L., E.I. and J.N. made the protein. S.F. processed the data. L.L.L. and J.N. critiqued the manuscript. J.A.M. and J.E.H. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Methods (PDF 4246 kb)
Rights and permissions
About this article
Cite this article
Mears, J., Lackner, L., Fang, S. et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18, 20–26 (2011). https://doi.org/10.1038/nsmb.1949
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1949
This article is cited by
-
OPA1 helical structures give perspective to mitochondrial dysfunction
Nature (2023)
-
Mitochondrial heterogeneity in diseases
Signal Transduction and Targeted Therapy (2023)
-
The role of mitochondria in rheumatic diseases
Nature Reviews Rheumatology (2022)
-
Selective inhibition of miRNA processing by a herpesvirus-encoded miRNA
Nature (2022)
-
Mitochondrial network remodeling: an important feature of myogenesis and skeletal muscle regeneration
Cellular and Molecular Life Sciences (2021)