Abstract
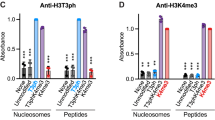
Phosphorylation of Ser10 of histone H3 regulates chromosome condensation and transcriptional activity. Using time-resolved, high-resolution NMR spectroscopy, we demonstrate that histone H3 Ser10 phosphorylation inhibits checkpoint kinase 1 (Chk1)– and protein kinase C (PKC)–mediated modification of Thr11 and Thr6, the respective primary substrate sites of these kinases. On unmodified H3, both enzymes also target Ser10 and thereby establish autoinhibitory feedback states on individual H3 tails. Whereas phosphorylated Ser10 does not affect acetylation of Lys14 by Gcn5, phosphorylated Thr11 impedes acetylation. Our observations reveal mechanistic hierarchies of H3 phosphorylation and acetylation events and provide a framework for intramolecular modification cross-talk within the N terminus of histone H3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Banerjee, T. & Chakravarti, D. A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 31, 4858–4873 (2011).
Baek, S.H. When signaling kinases meet histones and histone modifiers in the nucleus. Mol. Cell 42, 274–284 (2011).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Latham, J.A. & Dent, S.Y. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14, 1017–1024 (2007).
Davie, J.R. MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Sci. STKE 2003, PE33 (2003).
Wei, Y., Yu, L., Bowen, J., Gorovsky, M.A. & Allis, C.D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97, 99–109 (1999).
Metzger, E. et al. Phosphorylation of histone H3T6 by PKCβ(I) controls demethylation at histone H3K4. Nature 464, 792–796 (2010).
Garske, A.L. et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat. Chem. Biol. 6, 283–290 (2010).
Metzger, E. et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 10, 53–60 (2008).
Preuss, U., Landsberg, G. & Scheidtmann, K.H. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31, 878–885 (2003).
Shimada, M. et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 (2008).
Selenko, P. et al. In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 15, 321–329 (2008).
Hsu, J.Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000).
Lau, P.N. & Cheung, P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. USA 108, 2801–2806 (2011).
Liokatis, S., Dose, A., Schwarzer, D. & Selenko, P. Simultaneous detection of protein phosphorylation and acetylation by high-resolution NMR spectroscopy. J. Am. Chem. Soc. 132, 14704–14705 (2010).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Lo, W.S. et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926 (2000).
Fry, C.J., Shogren-Knaak, M.A. & Peterson, C.L. Histone H3 amino-terminal tail phosphorylation and acetylation: synergistic or independent transcriptional regulatory marks? Cold Spring Harb. Symp. Quant. Biol. 69, 219–226 (2004).
Shogren-Knaak, M.A., Fry, C.J. & Peterson, C.L. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J. Biol. Chem. 278, 15744–15748 (2003).
Clements, A. et al. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12, 461–473 (2003).
Nowak, S.J. & Corces, V.G. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20, 214–220 (2004).
Soloaga, A. et al. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22, 2788–2797 (2003).
Yamamoto, Y., Verma, U.N., Prajapati, S., Kwak, Y.T. & Gaynor, R.B. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423, 655–659 (2003).
Zippo, A., De Robertis, A., Serafini, R. & Oliviero, S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 (2007).
Sassone-Corsi, P. et al. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285, 886–891 (1999).
Wissmann, M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9, 347–353 (2007).
Ng, S.S. et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448, 87–91 (2007).
Theillet, F.X. et al. Site-specific mapping and time-resolved monitoring of Lysine methylation by high-resolution NMR spectroscopy. J. Am. Chem. Soc. 134, 7616–7619 (2012).
Ruthenburg, A.J., Li, H., Patel, D.J. & Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Ruthenburg, A.J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Liokatis, S., Dose, A., Schwarzer, D. & Selenko, P. Simultaneous detection of protein phosphorylation and acetylation by high-resolution NMR spectroscopy. J. Am. Chem. Soc. 132, 14704–14705 (2010).
Luger, K., Rechsteiner, T.J. & Richmond, T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999).
Lowary, P.T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Case, D.A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Chen, P. et al. The 1.7 Å crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100, 681–692 (2000).
Sessa, F. et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 (2005).
Lowe, E.D. et al. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16, 6646–6658 (1997).
Acknowledgements
We thank P. Schmieder and M. Beerbaum for excellent NMR infrastructure maintenance. A.S. and W.F. acknowledge funding by the Max Planck Gesellschaft (MBG). S.J.E. was supported by a Boehringer Ingelheim Fund (BIF) fellowship. F.-X.T. acknowledges support from the Association pour la Recherche contre le Cancer (ARC). D.S. and P.S. are supported by Emmy Noether research grants (SCHW1163/3-1 and SE1794/1-1) from the Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Contributions
S.L. conceived and performed NMR and biochemical experiments, analyzed the data and wrote the paper. A.S. and S.J.E. conceived and performed biochemical experiments. F.-X.T. performed computational modeling. R.K. performed peptide synthesis. B.v.R. performed structural modeling and visualization. D.S., C.D.A. and W.F. conceived biochemical experiments, discussed data and wrote parts of the manuscript. P.S. developed the project, supervised all experiments, analyzed the data, wrote the manuscript and made the figures. All authors discussed ideas and concepts, carefully read the manuscript and approved of the conclusions drawn therein.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Note. (PDF 16284 kb)
Rights and permissions
About this article
Cite this article
Liokatis, S., Stützer, A., Elsässer, S. et al. Phosphorylation of histone H3 Ser10 establishes a hierarchy for subsequent intramolecular modification events. Nat Struct Mol Biol 19, 819–823 (2012). https://doi.org/10.1038/nsmb.2310
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2310
This article is cited by
-
Hydroxamic acid-modified peptide microarrays for profiling isozyme-selective interactions and inhibition of histone deacetylases
Nature Communications (2021)
-
Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy
Clinical Epigenetics (2020)
-
Regulation of DNA damage-induced ATM activation by histone modifications
Genome Instability & Disease (2020)
-
Time-resolved NMR monitoring of tRNA maturation
Nature Communications (2019)
-
Co-delivery of paclitaxel and cetuximab by nanodiamond enhances mitotic catastrophe and tumor inhibition
Scientific Reports (2017)