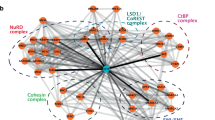
Abstract
During erythroid differentiation, β-globin gene expression is regulated by the locus control region (LCR). The transcription factor NF-E2p18/MafK binds within this region and is essential for β-globin expression in murine erythroleukemia (MEL) cells. Here we use the isotope-coded affinity tag (ICAT) technique of quantitative mass spectrometry to compare proteins interacting with NF-E2p18/MafK during differentiation. Our results define MafK as a 'dual-function' molecule that shifts from a repressive to an activating mode during erythroid differentiation. The exchange of MafK dimerization partner from Bach1 to NF-E2p45 is a key step in the switch from the repressed to the active state. This shift is associated with changes in the interaction of MafK with co-repressors and co-activators. Thus, our results suggest that in addition to its role as a cis-acting activator of β-globin gene expression in differentiated erythroid cells, the LCR also promotes an active repression of β-globin transcription in committed cells before terminal differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Forrester, W.C. et al. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 4, 1637–1649 (1990).
Fraser, P. & Grosveld, F. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 10, 361–365 (1998).
Epner, E. et al. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol. Cell 2, 447–455 (1998).
Reik, A. et al. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol. 18, 5992–6000 (1998).
Bender, M.A., Bulger, M., Close, J. & Groudine, M. β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol. Cell 5, 387–393 (2000).
Bulger, M. et al. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23, 5234–5244 (2003).
Orkin, S.H. Transcription factors and hematopoietic development. J. Biol. Chem. 270, 4955–4958 (1995).
Hardison, R. et al. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205, 73–94 (1997).
Armstrong, J.A. & Emerson, B.M. Transcription of chromatin: these are complex times. Curr. Opin. Genet. Dev. 8, 165–172 (1998).
Levings, P.P. & Bungert, J. The human β-globin locus control region. Eur. J. Biochem. 269, 1589–1599 (2002).
Liu, D. et al. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc. Natl. Acad. Sci. USA 89, 3899–3903 (1992).
Ney, P.A., Sorrentino, B.P., McDonagh, K.T. & Nienhuis, A.W. Tandem AP-1-binding sites within the human β-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev. 4, 993–1006 (1990).
Sorrentino, B.P., Ney, P.A. & Nienhuis, A.W. Localization and characterization of the DNase I-hypersensitive site II (HS II) enhancer. A critical regulatory element within the β-globin locus-activating region. Ann. NY Acad. Sci. 612, 141–151 (1990).
Talbot, D., Philipsen, S., Fraser, P. & Grosveld, F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 9, 2169–2177 (1990).
Gong, Q.H., McDowell, J.C. & Dean, A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the epsilon-globin gene in vivo by 5′ hypersensitive site 2 of the β-globin locus control region. Mol. Cell. Biol. 16, 6055–6064 (1996).
Strauss, E.C., Andrews, N.C., Higgs, D.R. & Orkin, S.H. In vivo footprinting of the human α-globin locus upstream regulatory element by guanine and adenine ligation-mediated polymerase chain reaction. Mol. Cell. Biol. 12, 2135–2142 (1992).
Moi, P. & Kan, Y.W. Synergistic enhancement of globin gene expression by activator protein-1-like proteins. Proc. Natl. Acad. Sci. USA 87, 9000–9004 (1990).
Moon, A.M. & Ley, T.J. Functional properties of the β-globin locus control region in K562 erythroleukemia cells. Blood 77, 2272–2284 (1991).
Mignotte, V., Wall, L., deBoer, E., Grosveld, F. & Romeo, P.H. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 17, 37–54 (1989).
Andrews, N.C., Erdjument-Bromage, H., Davidson, M.B., Tempst, P. & Orkin, S.H. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362, 722–728 (1993).
Andrews, N.C. et al. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl. Acad. Sci. USA 90, 11488–11492 (1993).
Igarashi, K. et al. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367, 568–572 (1994).
Andrews, N.C. The NF-E2 transcription factor. Int. J. Biochem. Cell Biol. 30, 429–432 (1998).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Tao, W.A. & Aebersold, R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr. Opin. Biotechnol. 14, 110–118 (2003).
Ranish, J.A. et al. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33, 349–355 (2003).
Kotkow, K.J. & Orkin, S.H. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol. 15, 4640–4647 (1995).
Igarashi, K., Itoh, K., Hayashi, N., Nishizawa, M. & Yamamoto, M. Conditional expression of the ubiquitous transcription factor MafK induces erythroleukemia cell differentiation. Proc. Natl. Acad. Sci. USA 92, 7445–7449 (1995).
Francastel, C., Poindessous-Jazat, V., Augery-Bourget, Y. & Robert-Lezenes, J. NF-E2p18/mafK is required in DMSO-induced differentiation of Friend erythroleukemia cells by enhancing NF-E2 activity. Leukemia 11, 273–280 (1997).
Motohashi, H., Shavit, J.A., Igarashi, K., Yamamoto, M. & Engel, J.D. The world according to Maf. Nucleic Acids Res. 25, 2953–2959 (1997).
Friend, C., Scher, W., Holland, J.G. & Sato, T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc. Natl. Acad. Sci. USA 68, 378–382 (1971).
Tsiftsoglou, A.S. & Wong, W. Molecular and cellular mechanisms of leukemic hemopoietic cell differentiation: an analysis of the Friend system. Anticancer Res. 5, 81–99 (1985).
Sawado, T., Igarashi, K. & Groudine, M. Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98, 10226–10231 (2001).
Dignam, J.D., Lebovitz, R.M. & Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Han, D.K., Eng, J., Zhou, H. & Aebersold, R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19, 946–951 (2001).
Eng, J.K., McCormack, A.L. & Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in protein databases. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Oyake, T. et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095 (1996).
Sawado, T., Halow, J., Bender, M.A. & Groudine, M. The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 17, 1009–1018 (2003).
Leach, K.M. et al. Characterization of the human β-globin downstream promoter region. Nucleic Acids Res. 31, 1292–1301 (2003).
Feng, Q. & Zhang, Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274, 269–290 (2003).
Knoepfler, P.S. & Eisenman, R.N. Sin meets NuRD and other tails of repression. Cell 99, 447–450 (1999).
Olave, I.A., Reck-Peterson, S.L. & Crabtree, G.R. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71, 755–781 (2002).
Armstrong, J.A., Bieker, J.J. & Emerson, B.M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95, 93–104 (1998).
Chi, T.H. et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195–199 (2002).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Ryan, R.F. et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19, 4366–4378 (1999).
Lechner, M.S., Begg, G.E., Speicher, D.W. & Rauscher, F.J. 3rd. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 20, 6449–6465 (2000).
Vassallo, M.F. & Tanese, N. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. USA 99, 5919–5924 (2002).
Kanemaki, M. et al. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274, 22437–22444 (1999).
Wood, M.A., McMahon, S.B. & Cole, M.D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5, 321–330 (2000).
Cheng, X., Reginato, M.J., Andrews, N.C. & Lazar, M.A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol. Cell. Biol. 17, 1407–1416 (1997).
Chen, C.J., Deng, Z., Kim, A.Y., Blobel, G.A. & Lieberman, P.M. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21, 476–487 (2001).
Wadman, I.A. et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145–3157 (1997).
Xu, Z., Huang, S., Chang, L.S., Agulnick, A.D. & Brandt, S.J. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23, 7585–7599 (2003).
Ge, H., Si, Y. & Roeder, R.G. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17, 6723–6729 (1998).
Westermarck, J. et al. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 21, 451–460 (2002).
Igarashi, K. et al. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 273, 11783–11790 (1998).
Yoshida, C. et al. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4, 643–655 (1999).
Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J.D. & Yamamoto, M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294, 1–12 (2002).
Johnson, K.D., Christensen, H.M., Zhao, B. & Bresnick, E.H. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8, 465–471 (2001).
Bulger, M., Sawado, T., Schubeler, D. & Groudine, M. ChIPs of the β-globin locus: unraveling gene regulation within an active domain. Curr. Opin. Genet. Dev. 12, 170–177 (2002).
Hung, H.L., Kim, A.Y., Hong, W., Radowski, C. & Blobel, G.A. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem. 276, 10715–10721 (2001).
Benezra, R., Cantor, C.R. & Axel, R. Nucleosomes are phased along the mouse β-major globin gene in erythroid and nonerythroid cells. Cell 44, 697–704 (1986).
Ogawa, K. et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 (2001).
Grandori, C., Cowley, S.M., James, L.P. & Eisenman, R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell. Dev. Biol. 16, 653–699 (2000).
Khavari, P.A., Peterson, C.L., Tamkun, J.W., Mendel, D.B. & Crabtree, G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174 (1993).
Nielsen, A.L. et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18, 6385–6395 (1999).
Griffin, T.J. et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 323–333 (2002).
Soutoglou, E. & Talianidis, I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295, 1901–1904 (2002).
Acknowledgements
We thank F.J. Dilworth and T. Sawado for helpful discussions, F.J. Dilworth for critically reading the manuscript, J. Eng and A. Nesvizhskii for help in the analysis of the mass spectrometry data, E. Soutoglou for helpful advice on the ChIP experiments, J. Sun for the generation of Bach1 antiserum, P. Chambon for the gift of the HP1γ antibody and V. Blank for the gift of the MafG antibody. M.B. is supported by a long-term fellowship from the Human Frontier Science Program Organization. This project has been funded in part with federal funds as part of the NHLBI Proteomics Initiative from the National Heart, Lung and Blood Institute, US National Institutes of Health (NIH), under contract no. N01-HV-28179 (R.A.), and with NIH grants DK44746 and HL57620 (M.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Brand, M., Ranish, J., Kummer, N. et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 11, 73–80 (2004). https://doi.org/10.1038/nsmb713
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb713
This article is cited by
-
BACH transcription factors in innate and adaptive immunity
Nature Reviews Immunology (2017)
-
Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells
Cellular and Molecular Life Sciences (2015)
-
Protein–protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches
Cellular and Molecular Life Sciences (2014)
-
ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors
Nature (2013)
-
Chromatin Loop Formation in the β-Globin Locus and Its Role in Globin Gene Transcription
Molecules and Cells (2012)