Abstract
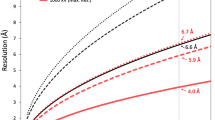
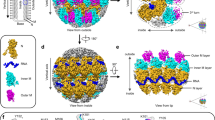
Biological membranes are notoriously resistant to structural analysis. Excellent candidates to tackle this problem in situ are membrane-containing viruses where the membrane is constrained by an icosahedral capsid. Cryo-EM and image reconstruction of bacteriophage PM2 revealed a membrane bilayer following the internal surface of the capsid. The viral genome closely interacts with the inner leaflet. The capsid, at a resolution of 8.4 Å, reveals 200 trimeric capsomers with a pseudo T = 21 dextro organization. Pentameric receptor-binding spikes protrude from the surface. It is evident from the structure that the PM2 membrane has at least two important roles in the life cycle. First, it acts as a scaffold to nucleate capsid assembly. Second, after host recognition, it fuses with the host outer membrane to promote genome entry. The structure also sheds light on how the viral supercoiled circular double-stranded DNA genome might be packaged and released.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Grahn, A.M., Daugelavicvius, R. & Bamford, D.H. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46, 1199–1209 (2002).
Mindich, L. Reverse genetics of dsRNA bacteriophage φ6. Adv. Virus Res. 53, 341–353 (1999).
Bamford, D.H., Caldentey, J. & Bamford, J.K. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45, 281–319 (1995).
Kivelä, H.M., Männistö, R.H., Kalkkinen, N. & Bamford, D.H. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262, 364–374 (1999).
Espejo, R.T. & Canelo, E.S. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34, 738–747 (1968).
Franklin, R.M., Hinnen, R., Schäfer, R. & Tsukagoshi, N. Structure and assembly of lipid-containing viruses, with special reference to bacteriophage PM2 as one type of model system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 276, 63–80 (1976).
Männistö, R.H., Kivelä, H.M., Paulin, L., Bamford, D.H. & Bamford, J.K.H. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262, 355–363 (1999).
Kivelä, H.M., Kalkkinen, N. & Bamford, D.H. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 76, 8169–8178 (2002).
Männistö, R.H., Grahn, A.M., Bamford, D.H. & Bamford, J.K.H. Transcription of bacteriophage PM2 involves phage-encoded regulators of heterologous origin. J.Bacteriol. 185, 3278–3287 (2003).
Tsukagoshi, N., Kania, M.N. & Franklin, R.M. Identification of acyl phosphatidylglycerol as a minor phospholipid of Pseudomonas BAL-31. Biochim. Biophys. Acta 450, 131–136 (1976).
Camerini-Otero, R.D. & Franklin, R.M. Structure and synthesis of a lipid-containing bacteriophage. XII. The fatty acids and lipid content of bacteriophage PM2. Virology 49, 385–393 (1972).
Brewer, G.J. Membrane-localized replication of bacteriophage PM2. Virology 84, 242–245 (1978).
Espejo, R.T., Canelo, E.S. & Sinsheimer, R.L. Replication of bacteriophage PM2 deoxyribonucleic acid: a closed circular double-stranded molecule. J. Mol. Biol. 56, 597–621 (1971).
Zimmer, S.G. & Millette, R.L. DNA-dependent RNA polymerase from Pseudomonas BAL-31. II. Transcription of the allomorphic forms of bacteriophage PM2 DNA. Biochemistry 14, 300–307 (1975).
Brewer, G.J. Control of membrane morphogenesis in bacteriophage. Int. Rev. Cytol. 68, 53–96 (1980).
Schäfer, R., Hinnen, R. & Franklin, R.M. Structure and synthesis of a lipid-containing bacteriophage. Properties of the structural proteins and distribution of the phospholipid. Eur. J. Biochem. 50, 15–27 (1974).
Baker, T.S., Olson, N.H. & Fuller, S.D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63, 862–922 (1999).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Harrison, S.C., Caspar, D.L., Camerini-Otero, R.D. & Franklin, R.M. Lipid and protein arrangement in bacteriophage PM2. Nat. New Biol. 229, 197–201 (1971).
Harauz, G. & van Heel, M. Similarity measures between images. Exact filters for general geometry of 3D reconstructions. Optik 73, 146–156 (1986).
Wikoff, W.R. et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 (2000).
Belnap, D.M., Olson, N.H. & Baker, T.S. A method for establishing the handedness of biological macromolecules. J. Struct. Biol. 120, 44–51 (1997).
Jiang, W., Baker, M.L., Ludtke, S.J. & Chiu, W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308, 1033–1044 (2001).
San Martín, C. et al. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 9, 756–763 (2002).
Mancini, E.J., Clarke, M., Gowen, B.E., Rutten, T. & Fuller, S.D. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5, 255–266 (2000).
Zhang, W. et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10, 907–912 (2003).
Kelley, L.A., MacCallum, R.M. & Sternberg, M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520 (2000).
Xu, L., Benson, S.D., Butcher, S.J., Bamford, D.H. & Burnett, R.M. The receptor-binding protein, P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure 11, 309–322 (2003).
Kivelä, H.M., Daugelavicvius, R., Hankkio, R.H., Bamford, J.K.H. & Bamford, D.H. Penetration of membrane-containing dsDNA bacteriophage PM2 into Pseudoalteromonas hosts. J. Bacteriol. 186 (in the press).
Tsukagoshi, N., Schäfer, R. & Franklin, R.M. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur. J. Biochem. 77, 585–588 (1977).
Earnshaw, W.C. & Casjens, S.R. DNA packaging by the double-stranded DNA bacteriophages. Cell 21, 319–331 (1980).
Gordon-Shaag, A., Ben-Nun-Shaul, O., Roitman, V., Yosef, Y. & Oppenheim, A. Cellular transcription factor Sp1 recruits Simian virus 40 capsid proteins to the viral packaging signal, ses. J. Virol. 76, 5915–5924 (2002).
Hayashi, M., Aoyama, A., Richardson, D.L. & Hayashi, M.N. Biology of the bacteriophage φX174. in The Bacteriophages, Vol. 2 (ed. Calendar, R.) 1–71 (Plenum Press, New York, 1988).
Smith, D.E. et al. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001).
Liddington, R.C. et al. Structure of simian virus 40 at 3.8-Å resolution. Nature 354, 278–284 (1991).
Stewart, P.L., Fuller, S.D. & Burnett, R.M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12, 2589–2599 (1993).
Rydman, P.S., Bamford, J.K.H. & Bamford, D.H. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 313, 785–795 (2001).
Grimes, J.M. et al. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5, 885–893 (1997).
Schägger, H. & von Jagow, G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 (1987).
Grigorieff, N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 277, 1033–1046 (1998).
Kivioja, T., Ravantti, J., Verkhovsky, A., Ukkonen, E. & Bamford, D. Local average intensity-based method for identifying spherical particles in electron micrographs. J.Struct. Biol. 131, 126–134 (2000).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Heymann, J.B. Bsoft: image and molecular processing in electron microscopy. J.Struct. Biol. 133, 156–169 (2001).
Fuller, S.D., Butcher, S.J., Cheng, R.H. & Baker, T.S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116, 48–55 (1996).
Baker, T.S. & Cheng, R.H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116, 120–130 (1996).
Ji, Y., Marinescu, D.C., Zhang, W. & Baker, T.S. Orientation refinement of virus structures with unknown symmetry. in Proceedings of the 17th International Parallel & Distributed Processing Symposium. (IEEE Press, Nice, France, 2003).
Marinescu, D.C., Ji, Y. & Lynch, R.E. Space-time tradeoffs for parallel 3D reconstruction algorithms for atomic virus structure determination. Concurrency Computation: Pract. Exper. 13, 1083–1106 (2001).
Huang, C.C., Couch, G.S., Pettersen, E.F. & Ferrin, T.E. Chimera: an extensible molecular modeling application constructed using standard components. Pacific Symposium on Biocomputing 1, 724 (1996).
Cheng, N. et al. Handedness of the herpes simplex virus capsid and procapsid. J.Virol. 76, 7855–7859 (2002).
Chacon, P. & Wriggers, W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317, 375–384 (2002).
Acknowledgements
We thank P. Laurinmäki for EM; L. Valmu of the Protein Chemistry core facility, Institute of Biotechnology, for MS analyses; R. Duda for the gift of HK97 proheads; D. Belnap, R. Burnett, J. Conway, S. Fuller, B. Heymann and Y. Ji for software and helpful discussions; Finnish IT Center for Science (CSC) for computer facilities. The work was funded by the National Graduate School in Informational and Structural Biology (J.T.H.), Helsinki Graduate School in Biotechnology and Molecular Biology (H.M.K.), the Academy of Finland Centre of Excellence Programme (2000–2005) (D.H.B.) and an Academy of Finland Research Fellowship (S.J.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Huiskonen, J., Kivelä, H., Bamford, D. et al. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol 11, 850–856 (2004). https://doi.org/10.1038/nsmb807
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb807
This article is cited by
-
Cryo-EM structure of ssDNA bacteriophage ΦCjT23 provides insight into early virus evolution
Nature Communications (2022)
-
The so far farthest reaches of the double jelly roll capsid protein fold
Virology Journal (2018)
-
Virus evolution: how far does the double β-barrel viral lineage extend?
Nature Reviews Microbiology (2008)