Abstract
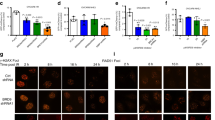
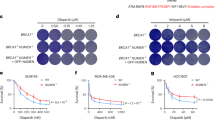
FOXM1 is implicated in genotoxic drug resistance but its role and mechanism of action remain unclear. Here, we establish that γH2AX foci, indicative of DNA double-strand breaks (DSBs), accumulate in a time-dependent manner in the drug-sensitive MCF-7 cells but not in the resistant counterparts in response to epirubicin. We find that FOXM1 expression is associated with epirubicin sensitivity and DSB repair. Ectopic expression of FOXM1 can increase cell viability and abrogate DSBs sustained by MCF-7 cells following epirubicin, owing to an enhancement in repair efficiency. Conversely, alkaline comet and γH2AX foci formation assays show that Foxm1-null cells are hypersensitive to DNA damage, epirubicin and γ-irradiation. Furthermore, we find that FOXM1 is required for DNA repair by homologous recombination (HR) but not non-homologous end joining (NHEJ), using HeLa cell lines harbouring an integrated direct repeat green fluorescent protein reporter for DSB repair. We also identify BRIP1 as a direct transcription target of FOXM1 by promoter analysis and chromatin-immunoprecipitation assay. In agreement, depletion of FOXM1 expression by small interfering RNA downregulates BRIP1 expression at the protein and mRNA levels in MCF-7 and the epirubicin-resistant MCF-7 EpiR cells. Remarkably, the requirement for FOXM1 for DSB repair can be circumvented by reintroduction of BRIP1, suggesting that BRIP1 is an important target of FOXM1 in DSB repair. Indeed, like FOXM1, BRIP1 is needed for HR. These data suggest that FOXM1 regulates BRIP1 expression to modulate epirubicin-induced DNA damage repair and drug resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lin SX, Chen J, Mazumdar M, Poirier D, Wang C, Azzi A et al. Molecular therapy of breast cancer: progress and future directions. Nat Rev Endocrinol 2010; 6: 485–493.
Alvarez RH, Valero V, Hortobagyi GN . Emerging targeted therapies for breast cancer. J Clin Oncol 2010; 28: 3366–3379.
Zelnak A . Overcoming taxane and anthracycline resistance. Breast J 2010; 16: 309–312.
Stearns V, Davidson NE, Flockhart DA . Pharmacogenetics in the treatment of breast cancer. Pharmacogenomics J 2004; 4: 143–153.
Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L . Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 2004; 56: 185–229.
Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF . Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 1984; 226: 466–468.
Raguz S, Yague E . Resistance to chemotherapy: new treatments and novel insights into an old problem. Br J Cancer 2008; 99: 387–391.
Myatt SS, Lam EW . The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007; 7: 847–859.
Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics 1997; 46: 435–442.
Yao KM, Sha M, Lu Z, Wong GG . Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem 1997; 272: 19827–19836.
Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008; 10: 1076–1082.
Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P . Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol 2008; 28: 5162–5171.
Karadedou CT, Gomes AR, Chen J, Petkovic M, Ho KK, Zwolinska AK et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene 2011; 31: 1845–1858.
Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res 2009; 69: 3501–3509.
Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res 2010; 8: 24–34.
Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat 2010; 122: 337–346.
Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene 2007; 26: 6212–6219.
Koo CY, Muir KW, Lam EW . FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta 2012; 1819: 28–37.
Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol 2006; 20: 1853–1866.
Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene 2010; 29: 2983–2995.
Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem 2006; 281: 25167–25176.
Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P . FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res 2010; 70: 5054–5063.
McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther 2009; 8: 582–591.
Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther 2011; 10: 1046–1058.
Tan Y, Raychaudhuri P, Costa RH . Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 2007; 27: 1007–1016.
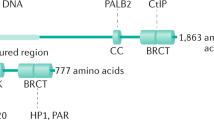
Cantor S . The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci 2004; 101: 2357–2362.
Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001; 105: 149–160.
Turnbull C, Rahman N . Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet [Review] 2008; 9: 321–345.
Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 2005; 8: 255–265.
Xie J, Litman R, Wang S, Peng M, Guillemette S, Rooney T et al. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene 2010; 29: 2499–2508.
Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmuller L . BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem J 2012; 441: 919–926.
Yu X, Chini CC, He M, Mer G, Chen J . The BRCT domain is a phospho-protein binding domain. Science 2003; 302: 639–642.
Delacote F, Han M, Stamato TD, Jasin M, Lopez BS . An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res 2002; 30: 3454–3463.
Weinstock DM, Brunet E, Jasin M . Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 2007; 9: 978–981.
Kruse V, Rottey S, De Backer O, Van Belle S, Cocquyt V, Denys H . PARP inhibitors in oncology: a new synthetic lethal approach to cancer therapy. Acta Clin Belg 2011; 66: 2–9.
de Olano N, Koo CY, Monteiro LJ, Pinto PH, Gomes AR, Aligue R et al. The p38 MAPK-MK2 axis regulates E2F1 and FOXM1 expression after epirubicin treatment. Mol Cancer Res 2012; 10: 1189–1202.
Palmieri C, Krell J, James CR, Harper-Wynne C, Misra V, Cleator S et al. Rechallenging with anthracyclines and taxanes in metastatic breast cancer. Nat Rev Clin Oncol [Review] 2010; 7: 561–574.
Park YY, Jung SY, Jennings N, Rodriguez-Aguayo C, Peng G, Lee SR et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis 2012; 33: 1843–1853.
Gong Z, Kim JE, Leung CC, Glover JN, Chen J . BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell 2010; 37: 438–446.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868.
Garinis GA, Mitchell JR, Moorhouse MJ, Hanada K, de Waard H, Vandeputte D et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J 2005; 24: 3952–3962.
Myatt SS, Lam EW . Targeting FOXM1. Nat Rev Cancer 2008; 8: 242.
Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S . The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem 2011; 3: 725–731.
Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 2005; 7: 126–136.
Bennardo N, Cheng A, Huang N, Stark JM . Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 2008; 4: e1000110.
Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M . Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol 2006; 409: 524–540.
Singh NP, McCoy MT, Tice RR, Schneider EL . A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 1988; 175: 184–191.
Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW . Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther 2008; 7: 2022–2032.
Nowak DE, Tian B, Brasier AR . Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 2005; 39: 715–725.
Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther 2010; 9: 844–855.
Acknowledgements
EW-FL, ARG and RCC were supported by grants from Cancer Research UK, EW-FL and C-YK by grants from Breast Cancer Campaign and Imperial College NHS Trust, LJM grants from Fundação para a Ciência e a Tecnologia, and PK and MK from the Royal Thai Government Scholarships. We also thank Dr Keng Heng for help with the microscopy and flow cytometry work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Monteiro, L., Khongkow, P., Kongsema, M. et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene 32, 4634–4645 (2013). https://doi.org/10.1038/onc.2012.491
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.491
Keywords
This article is cited by
-
KDM6 demethylases integrate DNA repair gene regulation and loss of KDM6A sensitizes human acute myeloid leukemia to PARP and BCL2 inhibition
Leukemia (2023)
-
The forkhead box M1 (FOXM1) expression and antitumor effect of FOXM1 inhibition in malignant rhabdoid tumor
Journal of Cancer Research and Clinical Oncology (2021)
-
Long noncoding RNA TRPM2-AS acts as a microRNA sponge of miR-612 to promote gastric cancer progression and radioresistance
Oncogenesis (2020)
-
FZD5 contributes to TNBC proliferation, DNA damage repair and stemness
Cell Death & Disease (2020)
-
Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer
Breast Cancer Research (2019)