Abstract
Increasing evidence supports the idea that cancer cell plasticity promotes metastasis and tumor recurrence, resulting in patient mortality. While it is clear that the tumor microenvironment (TME) contributes to cancer cell plasticity, the specific TME factors most actively controlling plasticity remain largely unknown. Here, we performed a screen to identify TME cytokines and growth factors that promote epithelial–mesenchymal plasticity, and acquisition of cancer stem cell (CSC) properties. Of 28 TME cytokines and growth factors tested, we identified Oncostatin M (OSM) as the most potent inducer of mesenchymal/CSC properties. OSM-induced plasticity was Signal Transducer and Activator of Transcription 3 (STAT3)-dependent, and also required a novel intersection with transforming growth factor-β (TGF-β)/SMAD signaling. OSM/STAT3 activation promoted SMAD3 nuclear accumulation, DNA binding and induced SMAD3-dependent transcriptional activity. Suppression of TGF-β receptor activity or ablation of SMAD3 or SMAD4, but not SMAD2, strongly suppressed OSM/STAT3-mediated plasticity. Moreover, removal of OSM or inhibition of STAT3 or SMAD3 resulted in a marked reversion to a non-invasive, epithelial phenotype. We propose that targeted blockade of the STAT3/SMAD3 axis in tumor cells may represent a novel therapeutic approach to prevent the plasticity required for metastatic progression and tumor recurrence.
Similar content being viewed by others
Introduction
Metastasis and tumor recurrence are the overwhelming causes of cancer-related mortality.1, 2 Yet, despite their importance, the molecular pathways involved in generating invasive, metastatic and therapy-resistant tumor cells remain poorly understood. Emerging evidence suggests that cancer cell plasticity contributes to tumor heterogeneity, which can promote metastasis and tumor recurrence.3, 4, 5 For example, epithelial–mesenchymal (E–M) plasticity is an important contributor to metastasis. Epithelial–mesenchymal transition (EMT) promotes the dissolution of tight cell–cell interactions by downregulation of epithelial proteins (claudin, occludin, ZO1, E-cadherin and cytokeratins).6, 7 The loss of cell–cell interactions is an important step in allowing escape from the primary tumor and entrance to the lymphatics or bloodstream. Importantly, metastatic tumors express epithelial markers resembling the primary tumor, suggesting that a reciprocal mesenchymal–epithelial transition occurs at the metastatic site.8, 9 In addition, circulating tumor cells are often enriched for mesenchymal markers, which correlates with therapeutic response.10, 11, 12, 13
We and others have shown that transformed human mammary epithelial cells (HMEC) acquire properties associated with breast cancer stem cell (CSC; identified by a CD24−/CD44+ cell surface marker profile) upon induction of EMT.14, 15, 16, 17, 18, 19, 20, 21 CSCs grow anchorage independently as tumorspheres, generate differentiated progeny and form xenografts that recapitulate patients’ primary tumors.22, 23 Likewise, tumor recurrence following therapy has been linked to cells harboring mesenchymal, CSC properties.5, 24, 25 In paired breast cancer core biopsies before and after chemotherapy, a higher percentage of CD24−/CD44+ CSC capable of forming tumorspheres exist following treatment, with the residual surviving cells harboring elevated CSC gene expression signatures.24 The enrichment of cells with a CSC phenotype following therapy has been interpreted in two different ways. The original interpretation posited that therapy kills non-CSC while sparing pre-existing CSC. However, evidence is emerging that supports another mechanism that non-CSC cancer cells can be induced into a CSC-like, drug-tolerant state by chemotherapy.19, 26, 27, 28, 29 A number of factors influence cancer cell plasticity, including intrinsic genetic and epigenetic changes, as well as extrinsic cues from the tumor microenvironment (TME). The TME is shaped, in part, by tumor-associated stromal cells, such as infiltrating immune cells, fibroblasts, adipocytes and endothelial cells, which are also corrupted during tumorigenesis.14, 30, 31, 32 Analysis of breast tumor stroma has identified altered levels of various cytokines, chemokines and growth factors emanating from the tumor stroma when compared to normal breast stroma.30, 33 However, while these TME factors are reported to contribute to cancer cell growth and survival, their influence over cancer cell plasticity remains unclear.
The current study assesses the ability of TME cytokines to cooperate with defined, intrinsic, genetic changes to generate cancer cell plasticity. One such cytokine, Oncostatin M (OSM), potently induced a Signal Transducer and Activator of Transcription 3 (STAT3)-dependent EMT, generating cells with CSC properties from purified epithelial/non-CSC populations. Importantly, we identified a link between STAT3 and the transforming growth factor-β (TGF-β) signaling effector SMAD3 that promoted OSM-induced EMT and generation of CSC. In essence, OSM receptor (OSMR) activation hijacks SMAD3-mediated transcription responsible for creating invasive, drug-tolerant CSC that facilitates metastatic progression and recurrence in epithelial/non-CSC populations. Our studies define how a novel OSM/STAT3/SMAD3 signaling axis promotes EMT and CSC expansion, and begin to explain why high levels of OSM within the TME may drive increased metastasis, tumor recurrence and ultimately poor patient prognosis.
Results
Cytokines aberrantly elevated in the TME promote cancer cell plasticity
Viral transduction of transforming genetic elements into primary HMEC results in the generation of two distinct, transformed cell populations. One transformed cell population retains epithelial characteristics (for example, E-cadherin expression) and exhibits a CD24+/CD44− cell surface marker profile, while another emergent population spontaneously acquires both mesenchymal characteristics (for example, Vimentin expression and spindle-shaped morphology) as well as a CD24−/CD44+ cell surface marker profile, indicative of breast CSC (Figure 1a).14 Fluorescence-activated cell sorting (FACS) for CD24+/CD44− or CD24−/CD44+ cells confirm that the CD24+/CD44− cells retain an epithelial character, while the CD24−/CD44+ cells have acquired a mesenchymal morphology (Figure 1b). Importantly, purified CD24−/CD44+ cells grow more efficiently in soft agar at limiting dilutions and as tumors that metastasize to the lung when orthotopically transplanted into immunocompromised mice.14 Moreover, exposure of the transformed epithelial/non-CSC (CD24+/CD44−) to exogenous TGF-β, a well-known driver of EMT, results in the acquisition of mesenchymal/CSC properties.14, 34, 35, 36, 37, 38 Thus, we postulated that our HMEC model provides a unique system to assess the impact of TME cytokines on the E–M and CSC plasticity governing aggressive cell properties.
Signals from the TME promote cellular plasticity. (a) Transformed HMEC consists of both epithelial/non-CSC (CD24+/CD44−) and mesenchymal/CSC (CD24−/CD44+) populations, that can be (b) separated by differential trypsinization or FACS. (c) Exogenous cytokine exposure drives cell-state changes of transformed HMEC (left) to either CD24−/CD44+ CSC (right, top) or CD24+/CD44− non-CSC (right, bottom). (d) Cytokine screen using a mixed population of transformed HMEC (as shown in a) that were exposed for 2 weeks to the indicated cytokines at the concentrations described in Table 1. Cytokine-treated populations were analyzed for CD24/CD44 using flow cytometry. Percent change in CSC was calculated as the difference in CSC population (identified as CD24−/CD44+, as shown in b) following exposure to each cytokine was calculated relative to untreated control.
A collection of 28 TME cytokines that have altered mRNA expression in invasive carcinoma (either micro-dissected stroma or tumor tissue) and are associated with one of three clinical breast cancer parameters, namely changes in (1) tumor recurrence; (2) metastasis; or (3) patient mortality, were identified (Table 1).30, 33, 39, 40 To test the impact of each TME cytokine on E–M and CSC plasticity, we exposed a mixed population of transformed HMEC to each of the 28 recombinant TME cytokines at the doses indicated in Table 1. Following 2 weeks of cytokine exposure, CD24 and CD44 cell surface marker expression was assessed by flow cytometry (Figure 1c). Nine of the cytokines (including TGF-β, used as a positive control) increased the percentage of CD24−/CD44+ CSC compared to untreated cells (Figure 1d). Interestingly, six of the nine cytokines that promoted CSC expansion are members of the interleukin-6 (IL-6) cytokine family and include IL-6, ciliary neurotrophic factor, cardiotrophin-1, cardiotrophin-like cytokine, leukemia inhibitory factor and OSM, with OSM providing the greatest expansion of mesenchymal/CSC, concomitant with the greatest level of STAT3 phosphorylation (Supplementary Figure S1). Conversely, a number of bone morphogenetic proteins (BMPs) and chemokine (C-X-C motif) ligand 16 (CXCL; among others) drove the expansion of the CD24+/CD44− non-CSC. These studies identify clinically important TME cytokines capable of altering the balance of epithelial/non-CSC and mesenchymal/CSC, and may help explain their association with more aggressive breast cancer phenotypes. Given the potency of OSM in driving mesenchymal/CSC expansion, it was chosen for further analysis.
Exogenous exposure of OSM induces EMT and acquisition of CSC properties
As designed, our TME cytokine screen could not distinguish whether transformed cells with mesenchymal/CSC properties were being created de novo by OSM, or whether a pre-existing population of mesenchymal/CSCs were selectively expanded. To test this, three populations of epithelial/non-CSCs were purified by FACS: all CD24+/CD44− (Figure 2a) and two subpopulations of CD24+/CD44− cells representing the highest CD24−expressing cells (CD24High/CD44−) or a moderate level of CD24 (CD24Medium/CD44−; Figure 2b). Each population was exposed to exogenous OSM immediately after sorting and assessed for changes in CD24 and CD44 cell surface marker expression over time. As early as 2 days after OSM addition, the emergence of a double-positive population became evident, followed by a reduction in CD24 expression days later (Figures 2a–c). A more rapid conversion was noted in the CD24Medium/CD44− population. Importantly, OSM did not affect the proliferation of either epithelial/non-CSC (CD24+/CD44−) or mesenchymal/CSC (CD24−/CD44+) populations (Figure 2d). In fact, mesenchymal/CSCs proliferate more slowly than the epithelial/non-CSC, arguing against the expansion of a pre-existing CSC population following OSM exposure. Moreover, whereas epithelial/non-CSC are arranged in a 'cobblestone' morphology identified by the presence of e-cadherin at the cell periphery, sustained exposure of non-CSC to OSM resulted in the progressive emergence of spindle-shaped, mesenchymal cells expressing vimentin and lacking E-cadherin (Figures 2e–g).
OSM exposure promotes EMT and the acquisition of CSC properties. (a) CD24+/CD44− and (b) CD24High/CD44− (top) or CD24Medium/CD44− (bottom) epithelial/non-CSCs were sorted by FACS, exposed to exogenous OSM and analyzed by flow cytometry for the emergence of CD24−/CD44+ CSC (% CSC shown in numbers) at each indicated time. (c) Quantification of the CD24−/CD44+ CSC from a, b. (d) FACS-sorted CD24+/CD44− non-CSCs and CD24−/CD44+ CSCs were exposed to OSM for 2, 3 and 4 days, and then growth assays assessed cell proliferation. (e–g) CD24+/CD44− epithelial/non-CSCs were exposed to OSM for 2 weeks and assessed by (e) brightfield microscopy (× 40), (f) confocal microscopy (× 100) for e-cadherin (red), vimentin (green) and nuclei (blue), or (g) western blot analysis for e-cadherin, vimentin and actin (as a loading control) protein levels.
Persistent OSM exposure is required to maintain mesenchymal/CSC properties
Breast CSCs are capable of anchorage-independent growth (AIG); therefore, we hypothesized that OSM would promote AIG of epithelial/non-CSC. Indeed, continuous exposure of epithelial/non-CSC populations to OSM resulted in efficient AIG relative to untreated cells in both the CD24High- and CD24Medium-sorted populations (Figure 3a). Interestingly, removal of OSM at the time of plating resulted in the loss of AIG capacity, suggesting that the maintenance of EMT and acquired CSC properties require continuous exposure to OSM (Figure 3a). Soft agar colonies generated by OSM treatment were recovered and further propagated in the presence or absence of OSM for an additional 2 or 4 weeks. Whereas recovered cells continuously exposed to OSM maintained a high proportion of cells with a CD24−/CD44+ profile (88.3%; Figure 3b), removal of OSM led to a majority of the CSC reverting back into CD24+/CD44− non-CSC. Similarly, epithelial/non-CSC continuously exposed to OSM in standard, two-dimensional tissue culture conditions (97.9% converted to CD24−/CD44+) also reverted back to an epithelial/non-CSC following OSM removal, even after sustained, long-term OSM exposure for 6 weeks. Again, a non-CSC population of cells with a 'cobblestone' morphology expressing E-cadherin at the periphery of cell clusters emerged from the CD44+, spindle-shaped mesenchymal cells upon OSM removal (Figures 3c–e). Taken together, our findings suggest that OSM can induce the acquisition of mesenchymal/CSC properties, such as AIG, and that sustained exposure to OSM is necessary for the maintenance of these aggressive phenotypes.
Maintenance of EMT and CSC properties requires sustained exposure to OSM. (a) AIG of CD24High/CD44− or CD24Medium/CD44− epithelial/non-CSC in soft agar after exposure to OSM for 2 weeks. At the time of plating, OSM was removed from cells that were previously exposed (removed). (b) CD24+/CD44− epithelial/non-CSC exposed to OSM and that grew anchorage independently were recovered from soft agar, grown in two-dimensional (2D) culture with or without continuous OSM exposure for another 2 or 4 weeks, and then analyzed by flow cytometry. % CSC depicted by indicated numbers. (c) CD24+/CD44− epithelial/non-CSCs were continuously exposed to OSM for 3 weeks, and then exposed to OSM or had OSM removed continuously for another 3 weeks (6 weeks total) and were assessed by flow cytometry. (d, e) CD24+/CD44− epithelial/non-CSCs were continuously exposed to OSM for 6 weeks, and then had OSM removed for 4 weeks and were analyzed by (d) brightfield microscopy (× 40), or (e) confocal microscopy (× 100) for e-cadherin (red), vimentin (green) and nuclei (blue).
OSM co-opts TGF-β/SMAD signaling to induce the acquisition of CSC properties
The cytokine TGF-β also induces CD24High/CD44− epithelial/non-CSC cells to acquire mesenchymal/CSC properties, and as observed with OSM, sustained TGF-β exposure is required to maintain the mesenchymal/CSC properties (Supplementary Figure S2A and S2B). In addition, our prior studies have defined a role for TGF-β signaling in the emergence of mesenchymal cells induced by oncogenic RAS.41 To assess whether TGF-β signaling contributes to OSM-mediated mesenchymal/CSC properties, epithelial/non-CSCs were treated for 2 weeks with OSM alone or in combination with the TGF-β receptor type I (TGFβRI) inhibitor SB431542. Whereas exposure to OSM resulted in the expected expansion of a CD24−/CD44+ mesenchymal/CSC population (from 1.7 to 33.8%), co-administration of SB431542 with OSM suppressed the generation of mesenchymal/CSC (Figures 4a and b). Since TGF-β-induced EMT is strongly associated with the invasiveness of mammary tumors,35, 36, 37, 42 we assessed whether OSM-induced EMT resulted in enhanced invasion, and whether SB431542 suppressed invasiveness in organotypic culture. Exposure of epithelial/non-CSC to OSM resulted in the emergence of invasive cells among the highly organized and densely packed spheroids (Figures 4c–e). Confocal microscopy determined that the untreated, non-invasive spheroids formed cell–cell interactions mediated by the presence of e-cadherin (red staining) in cells within the spheroids (Figure 4d). In contrast, vimentin expression (green staining) was observed in cells in the outer layer of the spheroids and in highly invasive cells no longer restricted by cell–cell interactions. Co-administration of SB431542 with OSM diminished the number of invasive spheroids, consistent with its ability to suppress EMT (Figure 4e). Taken together, these results suggest that EMT and acquisition of CSC properties can be suppressed by chemical inhibition of the TGF-β receptor, identifying a novel link between TGF-β signaling in OSM-mediated induction of EMT and acquisition of CSC properties.
Suppression of TGF-β signaling inhibits OSM-induced EMT, invasion and acquisition of CSC properties. CD24+/CD44− epithelial/non-CSC in (a, b) two-dimensional and (c–e) three-dimensional cultures were exposed for 2 weeks to OSM alone (no drug) or in combination with SB431542 (SB; 10 μM). Two-dimensional cultures were analyzed by (a) flow cytometry and (b) brightfield microscopy (× 4). (c–e) Three-dimensional organotypic cultures were analyzed by (c) brightfield microscopy (× 4); and (d) confocal microscopy (× 100) for e-cadherin (red), vimentin (green) and nuclei (blue); and (e) invasive spheroids per plate were quantified. (f, g) CD24+/CD44− epithelial/non-CSC (parental) expressing dominant-negative STAT3 (DNSTAT3) or TGF-β receptor II (DN-TGFβRII), SMAD7 and empty control vector (Vector) were (f) grown in 2D culture and analyzed by flow cytometry, or (g) were grown in 3D soft agar culture and assessed for AIG after exposure to exogenous OSM for 2 weeks.
The cooperation between OSM-induced EMT and acquisition of CSC properties, and TGF-β signaling is intriguing, yet many studies have reported the antagonistic actions of the downstream effectors STAT3 and SMADs on one another.43, 44, 45, 46 TGF-β ligand binds to TGF-β receptor type II (TGFβRII) and TGFβRI resulting in phosphorylation of TGFβRI and receptor-associated SMAD2 and SMAD3. SMAD2/3 complex with SMAD4 and translocate to the nucleus to affect gene expression. Transcriptional activity of SMAD2/4 or SMAD3/4 complexes is inhibited by SMAD7.47 To identify the components of TGF-β signaling required for OSM-induced EMT and acquisition of CSC phenotypes, epithelial/non-CSCs were infected with retroviruses encoding dominant-negative TGFβRII or SMAD7. Moreover, dominant-negative STAT3 (DNSTAT3) was used to confirm the importance of STAT3. All untreated derivatives retained an epithelial/non-CSC surface marker profile, similar to the parental population (Figure 4f). OSM resulted in expansion of the mesenchymal/CSC population to 42% in the vector control cells, which was efficiently blocked by DNSTAT3. Importantly, DNTGFβRII and SMAD7 also efficiently suppressed expansion of the mesenchymal/CSC population in response to OSM (Figure 4f). Similarly, DNSTAT3, DNTGFβRII- and SMAD7 inhibited OSM-induced AIG, consistent with the ability of each to suppress the expansion of CD24−/CD44+ CSC populations (Figure 4g). Taken together, these results suggest that OSM exposure engages TGFβR-mediated SMAD signaling to promote acquisition of CSC properties.
OSM/STAT3 utilizes SMAD3 to induce the acquisition of CSC properties
We next sought to determine which SMAD protein was responsible for OSM/STAT3-mediated acquisition of CSC properties. Epithelial/non-CSCs were infected with retroviruses encoding short hairpin RNA targeting SMAD2, SMAD3, SMAD4 or green fluorescent protein as a negative control and knockdown confirmed by western blot analysis (Figure 5a). Again, exposure of the control cells to OSM resulted in a robust expansion of the mesenchymal/CSC population (from 0.2 to 36.1%), which was inhibited by DNSTAT3 (Figure 5b). Interestingly, SMAD2 knockdown enhanced mesenchymal/CSC expansion (to 59.4%), whereas SMAD3 knockdown reduced the expansion of mesenchymal/CSC by greater than 70% (Figure 5b). SMAD4 reduced the acquisition of CSC properties less efficiently than SMAD3 knockdown (to 22%), likely due to the less efficient knockdown of SMAD4 protein (Figures 5a and b). These results suggest that OSM-induced EMT and acquisition of CSC properties require TGF-β signaling through a SMAD3/SMAD4 complex, specifically.
OSM-induced acquisition of CSC properties requires SMAD3. CD24+/CD44− epithelial/non-CSC expressing short hairpin RNA (shRNA) targeting SMAD2 (shSMAD2), SMAD3 (shSMAD3), SMAD4 (shSMAD4) or green fluorescent protein (shGFP) as a control were analyzed with control DNSTAT3 cells by (a) western blot analysis to confirm knockdown, or (b) flow cytometry before and after OSM exposure for 2 weeks. (c) CD24+/CD44− epithelial/non-CSCs were exposed to OSM or TGF-β for 3 weeks and analyzed by microarray hybridization. Gene expression was analyzed using GenomeStudio from Illumina identifying genes that changed in expression from untreated cells at least fivefold. The representation factor and P-value for overlapping expression were calculated using a Fisher’s exact test. (d) Transformed HMECs were plated in the presence and absence of either OSM or TGF-β1 for 0.5, 1 and 2 h. Western analysis (top) of whole-cell lysates assessed the proteins levels of Snail, phosphorylated-STAT3 Y705 (P-STAT3), STAT3, phosphorylated SMAD3 S423/425 (P-SMAD3), SMAD3 and Actin (as a loading control). Single-gene qRT-PCR analysis (bottom) using mRNA harvested from cells and primers targeting each specific gene assessed the fold-change expression levels of SNAI1, HAS2, SOCS3 and SERPINE1. (e) CD24+/CD44− cells were treated with OSM or TGF-β for 2 weeks, and western blot analysis for phosphorylated SMAD3 S423/425 (P-SMAD3), phosphorylated-SMAD2 S465/467 (P-SMAD2) and Actin (as a loading control) was performed.
Since SMAD3/SMAD4 complexes act as transcription factors, our results suggest that exposure to OSM should drive a SMAD3/SMAD4 transcriptional response similar to that induced by exposure to TGF-β. Epithelial/non-CSCs were exposed to OSM or TGF-β for 3 weeks and assessed for global gene expression changes compared to untreated cells. OSM exposure induced 1866 genes, while TGF-β exposure induced 851 genes by fivefold or more (Figure 5c). Importantly, 203 (24% of the TGF-β-induced genes) were activated similarly by OSM representing a 4.4-fold enrichment of overlap compared to random chance (P-value= 1.386 × 10–75). In addition, 2525 genes were repressed by OSM and 745 genes were repressed by TGF-β by at least fivefold, with 51% of the TGF-β repressed genes also being repressed by OSM. This represents a 6.6-fold enrichment of overlap compared to random chance (P-value=2.875 × 10–220). OSM and TGF-β exposure also induced highly similar changes in an EMT signature, a comparable emergence of a CD24LO/CD44HI CSC phenotype, and comparable AIG (Supplementary Figure S2C).48 The most straightforward explanation for the gene expression overlap is that OSM/STAT3 activity induces the autocrine activation of TGFβR-mediated signaling, thereby increasing SMAD3 phosphorylation, as described previously for IL-6.49 However, OSM did not increase TGFβR-mediated phosphorylation of either SMAD2 or SMAD3 proteins at a variety of time points after stimulation, ranging from 30 min (Figure 5d) to 14 days (Figure 5e). Despite the inability of OSM to increase SMAD3 phosphorylation, expression of the TGF-β-responsive gene encoding SNAIL, a key EMT transcription factor, was induced (both RNA and protein; Figure 5d). In addition, hyaluronan synthase 2 (HAS2), an enzyme responsible for the production of hyaluronic acid, which serves as a CD44 ligand to facilitate EMT,50 was also induced by OSM. Notably, the kinetics of SNAIL and HAS2 induction are distinct from direct STAT3 target gene SOCS3 and SMAD-target gene SERPINE1, in terms of both magnitude and timing of induction (Figure 5d). We found that the induction of Snail and P-STAT3 was more significant following exposure to OSM relative to the other IL-6 family cytokines tested in our screen, explaining the potency of OSM at inducing an EMT/CSC phenotype (Supplementary Figure S1).
To determine whether OSM-stimulated transactivation of SNAI1 and HAS2 requires TGF-β/SMAD3 signaling, epithelial/non-CSCs were treated for 2 h with OSM alone or in combination with the TGFβRI inhibitor SB431542 (10 μM) or the Janus Kinases 1 and 2 (JAK1/2) inhibitor Ruxolitinib (RUX; 10 μM), which prevents STAT3 phosphorylation in response to OSM. Exposure of epithelial/non-CSC to OSM induced an ~3.5-fold induction of SNAI1 and an ~5-fold induction of HAS2 (Figure 6a). OSM-induced SNAI1 and HAS2 expression was suppressed by TGFβRI inhibition, concomitant with reduced level of basal SMAD3 phosphorylation, both in whole-cell lysates and nuclear lysates (Figures 6a and b). SB431542 treatment did not have an impact on OSM-mediated STAT3 phosphorylation or expression of SOCS3, a direct STAT3 target gene. Inhibition of JAK1/2 using RUX prevented STAT3 phosphorylation and STAT3-mediated transcription of SNAI1, HAS2, as well as the direct target SOCS3 (Figure 6a). DNSTAT3 expression or short hairpin RNA-mediated ablation of SMAD3 and SMAD4 also suppressed OSM-mediated SNAIL induction, supporting our findings that STAT3 and SMAD3 are both required to promote a mesenchymal/CSC phenotype (Figures 6c, 5). Again, SMAD3/4 knockdown did not suppress OSM-mediated STAT3 phosphorylation or expression of SOCS3 (Figure 6c). Compared to other IL-6 family cytokines, OSM is unique in that it is a strong activator of STAT5 in addition to STAT3, which may implicate STAT5 in the OSM-induced phenotypes observed here. Yet, despite an immediate induction of STAT5 following exposure to OSM for 30 min, the activation of STAT5 was not maintained for an extended period of time (Supplementary Figure S3A and B). Moreover, DNSTAT3 expression, which strongly blocked OSM-induced mesenchymal/CSC properties, did not prevent STAT5 phosphorylation. In fact, STAT5 phosphorylation was more strongly activated in the DNSTAT3-expressing cells, whereas STAT3 phosphorylation and SNAIL induction were suppressed (Supplementary Figure S3C). These findings suggest that STAT5 activation is not a key determinant of the OSM-mediated phenotypes described here.
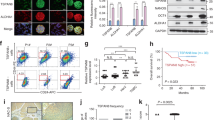
OSM-induced EMT and acquisition of CSC properties requires SMAD3 transcriptional activity in the nucleus. (a) CD24+/CD44− cells were exposed to OSM alone or in combination with RUX (10 μM) or SB431542 (SB; 10 μM) for 2 h, and then protein levels and gene expression were assessed by western blot analysis (top) and qRT-PCR analysis (bottom), respectively. (b) CD24+/CD44− cells were treated with SB431542 (SB) or TGF-β for 2 h and subjected to nuclear protein fractionation, and then western blot analysis of nuclear lysates for P-SMAD3 and Lamin B2 (as a loading control) was performed. (c) CD24+/CD44− epithelial/non-CSC expressing shGFP, shSMAD3, shSMAD4, or DNSTAT3 were exposed to OSM for 2 h, and then western blot analysis (top) assessed P-STAT3, STAT3, and Actin (as a loading control) protein levels; and qRT-PCR analysis (bottom) assessed SNAI1 and SOCS3 gene expression. (d) Chromatin immunoprecipitation (ChIP) and qRT-PCR analysis for SMAD3 enrichment on the SNAI1 and HAS2 gene promoters in CD24+/CD44− epithelial/non-CSC exposed to SB431542 for 24 h, and then treated with OSM alone, SB431542 alone or in combination for 1 h. (e) CD24+/CD44− epithelial/non-CSCs were exposed to OSM or TGF-β for 30 min, and then nuclear localization of SMAD3 (left) and STAT3 (right) was assessed by confocal microscopy (left panel) and quantified (right panel). Red triangles: cells with elevated nuclear localization of both SMAD3 and STAT3. Blue triangles: cells with low levels of SMAD3 and STAT3 nuclear localization (n=53 for the untreated and TGFβ-treated cells and n=74 for OSM-treated cells). (f, g) HPAC cells expressing SMAD7 or empty control Vector were treated with OSM for 7 days, and then (f) CD24/44 surface expression was determined via flow cytometry; in addition (g) protein levels of phosphorylated-STAT3 Y705 (P-STAT3), Snail and Actin (as a loading control) were determined by western blot analysis. (h) HPAC cells expressing shRNA targeting SMAD2 (S2), SMAD3 (S3) or GFP as a control were exposed to OSM for 7 days. Single-gene qRT-PCR analysis (top) assessed SNAI1 gene expression, and western blot analysis (bottom) confirmed SMAD2 and SMAD3 knockdown with Actin used as a loading control.
To assess whether STAT3 and SMAD3 proteins interact, a co-immunoprecipitation/western blot analysis was performed. Indeed, we found that STAT3 co-precipitated with SMAD3 (Supplementary Figure S4). In addition, we sought to determine whether OSM exposure enhanced the nuclear localization of SMAD3. Epithelial/non-CSCs were exposed to OSM, and nuclear localization of SMAD3 (or STAT3) was assessed by confocal microscopy. Cells treated with TGF-β were used as a positive control. As expected, OSM exposure drove the nuclear localization of STAT3 (Figure 6e). Interestingly, SMAD3 nuclear localization increased in response to OSM exposure, with the highest levels of nuclear SMAD3 present in cells with the highest level of nuclear STAT3 (Figure 6e, red triangles). Conversely, OSM-exposed cells with lower levels of nuclear STAT3 had lower levels of SMAD3 in the nucleus (Figure 6e, blue triangles). Finally, chromatin immunoprecipitation confirmed that OSM exposure enriched SMAD3 at both SNAI1 and HAS2 gene promoters (Figure 6d). Notably, this OSM-induced SMAD3 enrichment at both gene promoters was abrogated by inhibition of TGFβR1 with SB431542. Altogether, our results suggest that OSM-activated STAT3 interacts with SMAD3 to promote STAT3/SMAD3-mediated transcription of the SNAI1 and HAS2 genes. In addition to our HMEC model, we also assessed the OSM/STAT3/SMAD3 axis in a human pancreatic cancer cell line (HPAC cells), which undergo OSM-induced conversion to CD24−/CD44+ CSC (Figure 6f). Again, SMAD7 expression suppressed the emergence of a CD44+ population (from 67.5% in control cells to 24.4% in SMAD7-expressing cells; Figure 6f), concomitant with a reduction in Snail expression (Figure 6g). Furthermore, SMAD3 knockdown (but not SMAD2 knockdown) suppressed the OSM-mediated transcription of the Snail gene (Figure 6h).
Taken together, our data suggest that OSM exposure drives EMT and the generation of CSC by inducing a cooperative STAT3/SMAD3 gene transcription program. We propose that the OSMR/JAK-mediated phosphorylation of STAT3 induces its nuclear localization, which recruits basally phosphorylated SMAD3 to the promoters of select STAT3/SMAD3 targets (such as SNAIL and HAS2; Figure 7a). In contrast, if SMAD3 is unable to translocate to the nucleus because the TGFβR is inhibited or SMAD7 is expressed, then activated STAT3 is unable to cooperatively drive transcription of SNAIL, HAS2 or other STAT3/SMAD3-responsive genes, and EMT and CSC re-programming is prevented (Figure 7b).
Schematic of STAT3/SMAD3 signaling interactions. (a) Functional TGF-β signaling with OSM exposure drives a cooperative STAT3/SMAD3 gene transcription program. We propose that OSMR/JAK-mediated STAT3 activation recruits nuclear, basally phosphorylated SMAD3 to the promoters of select STAT3/SMAD3 targets. (b) Inactivation of TGF-β/SMAD3 signaling prevents SMAD3 from translocating to the nucleus and being recruited to specific promoters, thereby preventing the OSM/STAT3-mediated emergence of mesenchymal/CSC properties.
Discussion
Charles Darwin contended that, in the evolution of species, it is those most adaptable to change that survive and thrive, not the most innately intelligent or strong.51 Likewise, we contend that the same importance for adaptation exists in cancer biology, with increasing evidence suggesting that cancer cell plasticity is an important determinant of metastasis and tumor recurrence.4, 5, 14, 15, 24, 25, 26, 27, 28, 52, 53 Here, we assessed how TME cytokines found to be associated with patient outcomes influence E–M plasticity and CSC properties. One group of cytokines, including CXCL16, BMP7 and BMP10, promoted the expansion of a non-CSC population, consistent with previous reports that they induce mesenchymal–epithelial transition.54, 55, 56 In contrast, another group including hepatocyte growth factor and a number of IL-6 family members promoted the expansion of a mesenchymal/CSC phenotype, consistent with previous reports that they induce EMT.17, 57, 58 Of the 28 cytokines tested in our screen, OSM was the most potent inducer of E–M and CSC plasticity. Importantly, OSM did not merely select for mesenchymal/CSC, since they were generated from epithelial/non-CSC that were purified by FACS. Moreover, the dose of OSM used in our studies did not have an impact on either the growth of either the non-CSC or CSC populations. Our findings are consistent with a number of studies that argue that non-CSC can be converted into CSC via transitions between epithelial-like and mesenchymal-like states, by cytokines or chemotherapy.4, 5, 14, 15, 24, 25, 26, 27, 28, 52, 53 Taken together with the observation that OSM is significantly elevated in the breast, pancreatic, prostate, cervical and ovarian TME, the ability of OSM to promote cancer cell plasticity makes it a high priority for therapies capable of neutralizing its function.18, 59, 60, 61, 62, 63
OSM-induced EMT and generation of CSC was STAT3-dependent, suggesting that STAT3 activation induces the cell-state transitions that promote cancer cell plasticity. Indeed, most of the IL-6 family members, which signal through a common glycoprotein 130 (gp130) receptor subunit that activates STAT3, drove EMT and generation of CSC in our screen. The reason that OSM was particularly potent at inducing mesenchymal/CSC properties remains unclear, but may be due to the unique gp130/OSMR subunit β (OSMRβ) heterodimer (OSMR) complex. The OSMR complex has characteristics that are distinct from the gp130/gp130 homodimer receptor complexes that other IL-6 family-activate, and may account for the distinct signaling and biological effects of OSM relative to other IL-6 family members.64, 65, 66 For example, gp130/OSMRβ heterodimers can activate pathways not activated by other IL-6 family receptor complexes (STAT5, STAT6, AKT and PKC-δ), and also activate STAT3 more strongly.59, 67 OSMRβ has the unique capability of recruiting Src homology 2 domain-containing, which hyperactivates mitogen-activated protein kinase signaling.65 Interestingly, STAT3 serine 727 is phosphorylated in a MEK–ERK-dependent manner, and is required for RAS-mediated transformation.68 Thus, enhanced JAK-mediated tyrosine 705 phosphorylation and MEK/ERK-mediated serine 727 phosphorylation of STAT3 may explain the more robust effects of OSMR activation.
STAT3 can be activated by a number of additional signals beyond the OSM and other IL-6 family cytokines. A recent report demonstrated that oncogene-addicted tumor cells treated with epidermal growth factor receptor or MEK inhibitors activate STAT3 in response, which induced drug resistance, potentially by inducing E–M plasticity and CSC properties.69 Moreover, STAT3 can be activated by other oncogenic signaling cascades, including epidermal growth factor receptor, RAS, SRC, phosphatidylinositol 3-kinase and bone marrow X-linked non-receptor tyrosine kinase.68, 70, 71, 72, 73 Recently, a novel STAT3 inhibitor (BBI-608) was identified that selectively kills CD44+ CSC,74 with clinical trials currently underway to combine STAT3 inhibition with conventional chemotherapy in diverse cancer types. We propose that suppressing STAT3 activation, regardless of the mechanism of activation, may also be an effective strategy to block plasticity as a means to inhibit metastasis and tumor recurrence.75 Beyond JAK- and STAT3-specific inhibitors, we propose that targeting OSM in the TME or targeting the OSMR would also prevent the negative impact of STAT3 activation in cancer. Both OSM and OSMR have been targeted through use of neutralizing antibodies in models of arthritis and inflammatory heart failure, respectively.76, 77 Such neutralizing antibodies may be a beneficial approach to targeting OSM in the inflammatory TME to prevent the emergence of mesenchymal/CSC.
Beyond canonical JAK/STAT3 signaling, we also show that OSM-induced EMT and generation of CSC is dependent on TGF-β/SMAD3 signaling. OSM-induced activation of STAT3 promoted SMAD3 nuclear localization and transcriptional activation of STAT3/SMAD3 targets. The reason that SMAD3 is required for select OSM/STAT3 target genes is not fully clear. However, the ability of SMAD3 to bind to SMAD-binding elements to induce local chromatin remodeling will undoubtedly play a role. SMAD proteins bind to histone modifiers and histone readers such as JMJD3 and TRIM33, which can alter local chromatin structure and influence transcription.78, 79 We propose that nuclear chromatin-bound SMAD3 maintains the promoters of STAT3/SMAD3 target genes such as SNAIL and HAS2 in an open configuration, which is accessible to STAT3 upon its activation by OSM (Figure 7a). Conversely, the inhibition of basal SMAD3 phosphorylation would prevent its presence at STAT3/SMAD3 promoters, resulting in a closed chromatin confirmation that precludes STAT3 from binding following OSM exposure (Figure 7b). Since OSM expression can be induced by chemotherapy,80, 81 our identification of a novel link between STAT3 and SMAD3 transcriptional activity may explain, in part, the increase in TGF-β-responsive genes following treatment with chemotherapeutic agents.53
Our findings support the selective targeting of STAT3/SMAD3 target genes responsible for inducing E–M and CSC plasticity. A collective list of genes that are co-regulated by the STAT3/SMAD3 axis is currently unknown, as are the co-factors necessary for regulating their transcription, but subsequent studies will provide these important clues. Two genes examined here, SNAIL and HAS2, have known roles in EMT and CD44 regulation, and are likely important contributors to the observed phenotypes.50, 82, 83 Also unresolved are the disparate findings demonstrating that STAT3 and TGF-β/SMAD signaling can be both cooperative and antagonistic. For example, TGF-β signaling can potentiate IL-6/STAT3 activation, mediated by STAT3/SMAD3 complexes.84 In addition, JAK/STAT3 activity is required for TGF-β-induced Snail induction and EMT in RAS-driven tumor cells,85, 86, 87 albeit likely due to TGF-β-mediated IL-6 production, which activates canonical JAK/STAT3.88, 89, 90, 91, 92, 93 IL-6 exposure can activate SMAD3 to induce fibrosis, although this is due to elevated levels of SMAD3 phosphorylation, which is distinct from our observations.49 Such studies suggest that STAT3 and TGF-β/SMAD3 cooperatively engage one another; however, many studies have also identified antagonistic effects of the STAT3 and TGF-β/SMAD3 pathways on one another. For example, STAT3 activation can increase SMAD7 expression and form an inhibitory complex with SMAD3.43 Likewise, TGF-β signaling can prevent IL-6-mediated STAT3 activation, and thereby prevent its nuclear translocation and transcriptional activity.45, 46 The discrepancies among the studies of STAT3 and TGF-β/SMAD cross-talk may be rooted in the different tissue and cell types used in each study. The response of different cell types to cytokine stimulation is often cell-type- and context-specific, and is dependent on the genetic and epigenetic make-up of the cells. Identifying how TME cytokines like OSM have an impact on normal and transformed cells from the same tissue will help to define a therapeutic window as well as cancer-specific proteins and pathways that may serve as valuable new therapeutic targets.
Materials and methods
Cell growth and treatments
Specimen 48R HMEC were obtained from discarded surgical material under IRB approval, and grown as described previously.94, 95 HPAC cells were obtained from ATCC (Manassas, VA, USA, CRL-2119) and grown in DMEM/F12 supplemented with 10% fetal calf serum, 0.005 mg/ml of transferrin (#T2252; Sigma; St Louis, MO, USA), 10 ng/ml of EGF (#01-107; Millipore, Billerica, MA, USA), 0.002 mg/ml of Insulin (#I9278; Sigma) and 40 ng/ml of Hydrocortisone (#H4001; Sigma). HPAC cells were used from frozen stocks within 10 passages of receipt from ATCC. Cells were routinely tested for mycoplasma using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza, Basel, Switzerland). All cytokines and doses used in TME cytokine screen are described in Table 1. A single concentration of cytokine was administered to transformed 48 R HMEC over the course of 2 weeks before the cells were assessed for changes in CD24/CD44; the screen was performed a single time. Additional recombinant OSM (OSM10-13) or TGF-β (2001–2010) were obtained from DAPCEL Inc. (Cleveland, OH, USA) SB431542 was obtained from Sigma and RUX from LC Laboratories (Woburn, MA, USA). Retroviruses were generated and cells were transduced as previously described.41, 59, 95, 96, 97 Retroviral constructs encoding DNSTAT3, DNTGFβRII and SMAD7 are described previously.14, 59 The short hairpins pRetroSUPER targeting SMAD2 (15725), SMAD3 (15726) and SMAD4 (15727) were obtained from Addgene (Cambridge, MA, USA).98 Data shown are representative of three biological replicates using two independent cell models (48 R HMEC and HPAC).
Microscopy and flow cytometry
Brightfield and confocal images were captured as described previously.41 Three fields of each treatment were captured from three biological replicates, with one representative image presented in each figure. Quantification of nuclear localization was conducted using MetaMorph (Molecular Devices, Sunnyvale, CA, USA). Briefly, a region of interest (nucleus) was identified using DAPI, and the intensity of STAT3 and SMAD3 within the region of interest was calculated for untreated and treated cells at 30 min. A Mann–Whitney test (two-tailed) was used to assess statistically significant differences between the untreated, OSM- and TGFβ-treated groups. For flow cytometry, cell staining and analysis was conducted as described previously using CD24 (311106) and CD44 (338806) antibodies from BioLegend (San Diego, CA, USA).14 FACS was conducted on a BD Biosciences (San Jose, CA, USA) FACS ARIA II.
Western blot analysis and nuclear protein fraction isolation
Western blots were conducted as described previously.95 Representative blots are shown from three biological replicates. Antibodies used were e-cadherin (67A4), vimentin (V9) and Lamin B2 (F-8) from Santa Cruz Biotechnology (Dallas, TX, USA); SNAIL (C15D3), SMAD2 (D43B4), SMAD3 (C67H9), SMAD4 (9515), STAT3 (124H6), P-SMAD2 Ser465/467 (138D4), P-SMAD3 Ser423/425 (C25A9) and P-STAT3 Tyr705 (D3A7), P-STAT5 Y694 (D3A7) and STAT5 (9363) from Cell Signaling Technology (Danvers, MA, USA); moreover, ACTIN (ACTN05) is from Thermo Scientific (Waltham, MA, USA). For nuclear protein localization assays, cells were treated with SB431542 (10 μM) or TGF-β for 2 h and subjected to a subcellular protein fractionation kit (Thermo Scientific, #78840) according to the manufacturer’s instructions. Western blot analysis of nuclear-soluble protein fractions then examined P-SMAD3 and Lamin B2 protein nuclear localization.
Growth assays and three-dimensional cell cultures
For growth assays, cells were plated in the presence or absence of OSM in triplicate, technical replicates in six-well plates at 25 000 cells/well for the indicated days, and then total cell number for each well was quantified using a Beckman Coulter counter. Average total cell number per well for each triplicate at each indicated time is shown. For three-dimensional cultures, soft agar assays were described previously.41, 95 Error bars for growth assays and AIG represent s.d. The data presented are representative of three independent, biological replicates. Organotypic culture was described previously;59 data presented are one of three biological replicates.
Quantitative real-time PCR and gene expression analysis
Cells were pre-treated for 1h with pharmacologic inhibitors, and then treated for 2 h with OSM or TGF-β. Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Frederick, MD, USA) with on column DNaseI digest. One microgram RNA was reverse-transcribed using the iScript cDNA synthesis Kit (Bio-Rad, Hercules, CA, USA). Quantitative real-time PCR (qRT-PCR) was conducted with iQ SYBR Green Supermix (Bio-Rad) on a CFX96 thermocycler (Bio-Rad) in triplicate (technical replicates) to amplify genes using the primer sequences below using a 60 oC annealing temperature. Error bars for qRT-PCR represent s.d.; the data presented are representative of three independent, biological replicates. qRT-PCR primers: SNAI1 primers: Forward 5′-GGAAGCCTAACTACAGCGAG-3′ and Reverse 5′-GAGAGTCCCAGATGAGCATTG-3′; SERPINE1 primers: Forward 5′-CAGACCAAGAGCCTCTCCAC-3′ and Reverse 5′-ATCACTTGGCCCATGAAAAG-3′; HAS2 primers: Forward 5′-GGAAAGCTTGATTCAGACACTATGC-3′ and Reverse 5′-AGGGAATTCGTACAGCCATTCTCGG-3′; SOCS3 primers: Forward 5′-CAAGCACAAGAAGCCAAC-3′ and Reverse 5′-TTCCCTCCAACACATTCC-3′; and ACTIN primers (as a control): Forward 5′-CAGCCATGTACGTTGCTATCCAGG-3′ and Reverse 5′-AGGTCCAGACGCAGGATGGCATG. Error bars represent s.d. For gene expression analysis, RNA from one sample set was hybridized to a HumanHt-12 v4 Expression BeadChip (BD-103-0204, Illumina, San Diego, CA, USA) and analyzed using GenomeStudio from Illumina. Microarray data were deposited into the NCBI Gene Expression Omnibus (GEO) public functional genomics data repository (Accession number is GSE92297). Genes changed at least fivefold in expression were included in the analysis. The representation factor and P-value for overlapping expression were calculated using a Fisher’s exact test.
References
Nguyen DX, Bos PD, Massague J . Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 2009; 9: 274–284.
Weiss RB, Woolf SH, Demakos E, Holland JF, Berry DA, Falkson G et al. Natural history of more than 20 years of node-positive primary breast carcinoma treated with cyclophosphamide, methotrexate, and fluorouracil-based adjuvant chemotherapy: a study by the Cancer and Leukemia Group B. J Clin Oncol 2003; 21: 1825–1835.
Meacham CE, Morrison SJ . Tumour heterogeneity and cancer cell plasticity. Nature 2013; 501: 328–337.
Brooks MD, Burness ML, Wicha MS . Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 2015; 17: 260–271.
Doherty MR, Smigiel JM, Junk DJ, Jackson MW . Cancer stem cell plasticity drives therapeutic resistance. Cancers 2016; 8: E8.
Bastid J . EMT in carcinoma progression and dissemination: facts, unanswered questions, and clinical considerations. Cancer Metastasis Rev 2012; 31: 277–283.
Yang J, Weinberg RA . Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14: 818–829.
Gunasinghe NP, Wells A, Thompson EW, Hugo HJ . Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 2012; 31: 469–478.
Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015; 526: 131–135.
Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013; 339: 580–584.
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S . Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 2009; 11: R46.
Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani ME, Ow SG et al. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 2015; 6: 15578–15593.
Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, Nicolazzo C et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat 2011; 130: 449–455.
Junk DJ, Cipriano R, Bryson BL, Gilmore HL, Jackson MW . Tumor microenvironmental signaling elicits epithelial-mesenchymal plasticity through cooperation with transforming genetic events. Neoplasia 2013; 15: 1100–1109.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A . Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008; 3: e2888.
Xie G, Yao Q, Liu Y, Du S, Liu A, Guo Z et al. IL-6-induced epithelial-mesenchymal transition promotes the generation of breast cancer stem-like cells analogous to mammosphere cultures. Int J Oncol 2012; 40: 1171–1179.
West NR, Murray JI, Watson PH . Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 2014; 33: 1485–1494.
Wrighton KH, Lin X, Feng XH . Phospho-control of TGF-beta superfamily signaling. Cell Res 2009; 19: 8–20.
Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013; 154: 61–74.
Hollier BG, Tinnirello AA, Werden SJ, Evans KW, Taube JH, Sarkar TR et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res 2013; 73: 1981–1992.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003; 100: 3983–3988.
Rycaj K, Tang DG . Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Res 2015; 75: 4003–4011.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 2009; 106: 13820–13825.
Adorno-Cruz V, Kibria G, Liu X, Doherty M, Junk DJ, Guan D et al. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res 2015; 75: 924–929.
Goldman A, Majumder B, Dhawan A, Ravi S, Goldman D, Kohandel M et al. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun 2015; 6: 6139.
Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res 2011; 71: 5317–5326.
Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene 2012; 31: 432–445.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80.
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008; 14: 518–527.
Mbeunkui F, Johann DJ Jr . Cancer and the tumor microenvironment: a review of an essential relationship. Cancer Chemother Pharmacol 2009; 63: 571–582.
Egeblad M, Nakasone ES, Werb Z . Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010; 18: 884–901.
Celis JE, Gromov P, Cabezon T, Moreira JM, Ambartsumian N, Sandelin K et al. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics 2004; 3: 327–344.
Wendt MK, Allington TM, Schiemann WP . Mechanisms of the epithelial-mesenchymal transition by TGF-beta. Future Oncol 2009; 5: 1145–1168.
Heldin CH, Vanlandewijck M, Moustakas A . Regulation of EMT by TGFbeta in cancer. FEBS Lett 2012; 586: 1959–1970.
Morrison CD, Parvani JG, Schiemann WP . The relevance of the TGF-beta Paradox to EMT-MET programs. Cancer Lett 2013; 341: 30–40.
Derynck R, Muthusamy BP, Saeteurn KY . Signaling pathway cooperation in TGF-beta-induced epithelial-mesenchymal transition. Curr Opin Cell Biol 2014; 31: 56–66.
Xu J, Lamouille S, Derynck R . TGF-beta-induced epithelial to mesenchymal transition. Cell Res 2009; 19: 156–172.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6: 1–6.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352.
Cipriano R, Kan CE, Graham J, Danielpour D, Stampfer M, Jackson MW . TGF-beta signaling engages an ATM-CHK2-p53-independent RAS-induced senescence and prevents malignant transformation in human mammary epithelial cells. Proc Natl Acad Sci USA 2011; 108: 8668–8673.
Wendt MK, Schiemann BJ, Parvani JG, Lee YH, Kang Y, Schiemann WP . TGF-beta stimulates Pyk2 expression as part of an epithelial-mesenchymal transition program required for metastatic outgrowth of breast cancer. Oncogene 2013; 32: 2005–2015.
Luwor RB, Baradaran B, Taylor LE, Iaria J, Nheu TV, Amiry N et al. Targeting Stat3 and Smad7 to restore TGF-beta cytostatic regulation of tumor cells in vitro and in vivo. Oncogene 2013; 32: 2433–2441.
Wang G, Yu Y, Sun C, Liu T, Liang T, Zhan L et al. STAT3 selectively interacts with Smad3 to antagonize TGF-beta. Oncogene 2016; 35: 4388–4398.
Zhao S, Venkatasubbarao K, Lazor JW, Sperry J, Jin C, Cao L et al. Inhibition of STAT3 Tyr705 phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Res 2008; 68: 4221–4228.
Walia B, Wang L, Merlin D, Sitaraman SV . TGF-beta down-regulates IL-6 signaling in intestinal epithelial cells: critical role of SMAD-2. FASEB J 2003; 17: 2130–2132.
Luo L, Li N, Lv N, Huang D . SMAD7: a timer of tumor progression targeting TGF-beta signaling. Tumour Biol 2014; 35: 8379–8385.
Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA 2010; 107: 15449–15454.
O'Reilly S, Ciechomska M, Cant R, van Laar JM . Interleukin-6 (IL-6) trans signaling drives a STAT3-dependent pathway that leads to hyperactive transforming growth factor-beta (TGF-beta) signaling promoting SMAD3 activation and fibrosis via Gremlin protein. J Biol Chem 2014; 289: 9952–9960.
Heldin P, Basu K, Kozlova I, Porsch H . HAS2 and CD44 in breast tumorigenesis. Adv Cancer Res 2014; 123: 211–229.
Boero F . From Darwin's Origin of Species toward a theory of natural history. F1000prime Rep 2015; 7: 49.
Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008; 100: 672–679.
Bhola NE, Balko JM, Dugger TC, Kuba MG, Sanchez V, Sanders M et al. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest 2013; 123: 1348–1358.
Buijs JT, van der Horst G, van den Hoogen C, Cheung H, de Rooij B, Kroon J et al. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene 2012; 31: 2164–2174.
Fang Y, Henderson Jr FC, Yi Q, Lei Q, Li Y, Chen N . Chemokine CXCL16 expression suppresses migration and invasiveness and induces apoptosis in breast cancer cells. Mediators Inflamm 2014; 2014: 478641.
Ye L, Bokobza S, Li J, Moazzam M, Chen J, Mansel RE et al. Bone morphogenetic protein-10 (BMP-10) inhibits aggressiveness of breast cancer cells and correlates with poor prognosis in breast cancer. Cancer Sci 2010; 101: 2137–2144.
Farrell J, Kelly C, Rauch J, Kida K, Garcia-Munoz A, Monsefi N et al. HGF induces epithelial-to-mesenchymal transition by modulating the mammalian hippo/MST2 and ISG15 pathways. J Proteome Res 2014; 13: 2874–2886.
Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N et al. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009; 28: 2940–2947.
Kan CE, Cipriano R, Jackson MW . c-MYC functions as a molecular switch to alter the response of human mammary epithelial cells to oncostatin M. Cancer Res 2011; 71: 6930–6939.
West NR, Murphy LC, Watson PH . Oncostatin M suppresses oestrogen receptor-alpha expression and is associated with poor outcome in human breast cancer. Endocr Relat Cancer 2012; 19: 181–195.
Vlaicu P, Mertins P, Mayr T, Widschwendter P, Ataseven B, Hogel B et al. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer 2013; 13: 197.
Smith DA, Kiba A, Zong Y, Witte ON . Interleukin-6 and oncostatin-M synergize with the PI3K/AKT pathway to promote aggressive prostate malignancy in mouse and human tissues. Mol Cancer Res 2013; 11: 1159–1165.
Zhu M, Che Q, Liao Y, Wang H, Wang J, Chen Z et al. Oncostatin M activates STAT3 to promote endometrial cancer invasion and angiogenesis. Oncol Rep 2015; 34: 129–138.
Underhill-Day N, Heath JK . Oncostatin M (OSM) cytostasis of breast tumor cells: characterization of an OSM receptor beta-specific kernel. Cancer Res 2006; 66: 10891–10901.
Hermanns HM, Radtke S, Schaper F, Heinrich PC, Behrmann I . Non-redundant signal transduction of interleukin-6-type cytokines. The adapter protein Shc is specifically recruited to rhe oncostatin M receptor. J Biol Chem 2000; 275: 40742–40748.
Hermanns HM . Oncostatin M and interleukin-31: Cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev 2015; 26: 545–558.
Richards CD . The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm 2013; 2013: 512103.
Gough DJ, Koetz L, Levy DE . The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS ONE 2013; 8: e83395.
Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J . Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 2014; 26: 207–221.
Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell 2011; 19: 498–511.
Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR . STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci USA 2013; 110: 16975–16980.
Garcia R, Yu CL, Hudnall A, Catlett R, Nelson KL, Smithgall T et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ 1997; 8: 1267–1276.
Hart JR, Liao L, Yates 3rd JR, Vogt PK . Essential role of Stat3 in PI3K-induced oncogenic transformation. Proc Natl Acad Sci USA 2011; 108: 13247–13252.
Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci USA 2015; 112: 1839–1844.
Junk DJ, Bryson BL, Jackson MW . HiJAK'd signaling; the STAT3 paradox in senescence and cancer progression. Cancers 2014; 6: 741–755.
Plater-Zyberk C, Buckton J, Thompson S, Spaull J, Zanders E, Papworth J et al. Amelioration of arthritis in two murine models using antibodies to oncostatin M. Arthritis Rheum 2001; 44: 2697–2702.
Poling J, Gajawada P, Richter M, Lorchner H, Polyakova V, Kostin S et al. Therapeutic targeting of the oncostatin M receptor-beta prevents inflammatory heart failure. Basic Res Cardiol 2014; 109: 396.
Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA . H3K27me3 regulates BMP activity in developing spinal cord. Development 2010; 137: 2915–2925.
Dahle O, Kumar A, Kuehn MR . Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 2010; 3: ra48.
Singh RA, Sodhi A . Cisplatin-treated macrophages produce oncostatin M: regulation by serine/threonine and protein tyrosine kinases/phosphatases and Ca2+/calmodulin. Immunol Lett 1998; 62: 159–164.
Sodhi A, Shishodia S, Shrivastava A . Cisplatin-stimulated murine bone marrow-derived macrophages secrete oncostatin M. Immunol Cell Biol 1997; 75: 492–496.
Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S et al. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep 2014; 3: 691–698.
Baulida J, Garcia de Herreros A . Snail1-driven plasticity of epithelial and mesenchymal cells sustains cancer malignancy. Biochim Biophys Acta 2015; 1856: 55–61.
Yamamoto T, Matsuda T, Muraguchi A, Miyazono K, Kawabata M . Cross-talk between IL-6 and TGF-beta signaling in hepatoma cells. FEBS Lett 2001; 492: 247–253.
Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z et al. JAK/STAT3 signaling is required for TGF-beta-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol 2014; 44: 1643–1651.
Xu MY, Hu JJ, Shen J, Wang ML, Zhang QQ, Qu Y et al. Stat3 signaling activation crosslinking of TGF-beta1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim Biophys Acta 2014; 1842: 2237–2245.
Saitoh M, Endo K, Furuya S, Minami M, Fukasawa A, Imamura T et al. STAT3 integrates cooperative Ras and TGF-beta signals that induce Snail expression. Oncogene 2016; 35: 1049–1057.
Moon SU, Kang MH, Sung JH, Kim JW, Lee JO, Kim YJ et al. Effect of Smad3/4 on chemotherapeutic drug sensitivity in colorectal cancer cells. Oncol Rep 2015; 33: 185–192.
Turner M, Chantry D, Feldmann M . Transforming growth factor beta induces the production of interleukin 6 by human peripheral blood mononuclear cells. Cytokine 1990; 2: 211–216.
Seong GJ, Hong S, Jung SA, Lee JJ, Lim E, Kim SJ et al. TGF-beta-induced interleukin-6 participates in transdifferentiation of human Tenon's fibroblasts to myofibroblasts. Mol Vis 2009; 15: 2123–2128.
Eickelberg O, Pansky A, Mussmann R, Bihl M, Tamm M, Hildebrand P et al. Transforming growth factor-beta1 induces interleukin-6 expression via activating protein-1 consisting of JunD homodimers in primary human lung fibroblasts. J Biol Chem 1999; 274: 12933–12938.
Elias JA, Lentz V, Cummings PJ . Transforming growth factor-beta regulation of IL-6 production by unstimulated and IL-1-stimulated human fibroblasts. J Immunol 1991; 146: 3437–3443.
Park JI, Lee MG, Cho K, Park BJ, Chae KS, Byun DS et al. Transforming growth factor-beta1 activates interleukin-6 expression in prostate cancer cells through the synergistic collaboration of the Smad2, p38-NF-kappaB, JNK, and Ras signaling pathways. Oncogene 2003; 22: 4314–4332.
Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res 2009; 69: 7557–7568.
Junk DJ, Cipriano R, Stampfer M, Jackson MW . Constitutive CCND1/CDK2 activity substitutes for p53 loss, or MYC or oncogenic RAS expression in the transformation of human mammary epithelial cells. PLoS ONE 2013; 8: e53776.
Wang H, Song K, Krebs TL, Yang J, Danielpour D . Smad7 is inactivated through a direct physical interaction with the LIM protein Hic-5/ARA55. Oncogene 2008; 27: 6791–6805.
Tang B, de Castro K, Barnes HE, Parks WT, Stewart L, Bottinger EP et al. Loss of responsiveness to transforming growth factor beta induces malignant transformation of nontumorigenic rat prostate epithelial cells. Cancer Res 1999; 59: 4834–4842.
He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J . Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 2006; 125: 929–941.
Acknowledgements
DJJ was supported by NIH T32 CA059366. MWJ is supported by the US National Institutes of Health (R01CA138421) and the American Cancer Society (Research Scholar Award # RSG CCG-122517). Additional support was provided by the Case Comprehensive Cancer Center (P30 CA43703; Cytometry & Imaging Microscopy).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Junk, D., Bryson, B., Smigiel, J. et al. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 36, 4001–4013 (2017). https://doi.org/10.1038/onc.2017.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.33
This article is cited by
-
Cancer cell plasticity: from cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance
Cancer and Metastasis Reviews (2024)
-
Regulation and signaling pathways in cancer stem cells: implications for targeted therapy for cancer
Molecular Cancer (2023)
-
Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review
Signal Transduction and Targeted Therapy (2023)
-
The dynamic tumor–stromal crosstalk: implications of ‘stromal-hot’ tumors in the process of epithelial–mesenchymal transition in breast cancer
Molecular Biology Reports (2023)
-
Mechanistic insights into the interplays between neutrophils and other immune cells in cancer development and progression
Cancer and Metastasis Reviews (2022)