Abstract
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is known to have a critical role in DNA double-strand break repair. We have previously reported that DNA-PKcs is activated when cells enter mitosis and functions in mitotic spindle assembly and chromosome segregation. Here we report that DNA-PKcs is the upstream regulator of the Chk2–Brca1 pathway, which impacts microtubule dynamics, kinetochore attachment and chromosomal segregation in mitosis. Downstream from Chk2, Brca1 promotes monoubiquitination of γ-tubulin to inhibit microtubule nucleation and growth. We found that DNA-PKcs is essential for mitotic Chk2 phosphorylation at Thr68. As in Chk2- and Brca1-deficient cells, loss of DNA-PKcs resulted in chromosome misalignment and lagging during anaphase owing to elevation in microtubule dynamics. Importantly, these mitotic aberrations in DNA-PKcs-defective cells were alleviated by the overexpression of phosphomimetic Chk2 or Brca1 mutant proteins but not their wild-type counterparts. Taken together, these results demonstrate that DNA-PKcs regulates mitotic spindle organization and chromosomal instability via the Chk2–Brca1 signaling pathway.
Similar content being viewed by others
Introduction
Chromosome instability (CIN), characterized by increased frequency of gain or loss of the whole chromosomes, has long been implicated in tumorigenesis, poor patient prognosis and drug resistance.1 The most common cause of CIN in cancer cells is a defect in the dynamics of kinetochore-microtubule (k-MT) attachment during mitosis.2 An evolutionarily conserved spindle assembly checkpoint mechanism has evolved to ensure proper completion of all k-MT attachments before chromosomal segregation.3 Certain improper k-MT attachments (for example, merotelic attachment with microtubules orientated from both spindle poles) can escape from spindle assembly checkpoint surveillance and persist into the lagging chromosome; these are observed at the spindle midzone during anaphase.2 Abnormal k-MT attachments occur frequently in early mitosis but are efficiently corrected in normal cells.4 In cancer cells with CIN, however, stability of k-MT attachments increases, hindering the ability to correct the abnormal k-MT attachment.5 In addition, abnormal spindle assembly with extra centrosomes significantly increases the frequency of lagging chromosomes during anaphase, indicating that centrosome instability is also a major source of abnormal k-MT attachment.6 Centrosome hypertrophy is an accepted cause of CIN and is frequently correlated with tumorigenesis.7 Moreover, centrosome instability is highly linked to certain tumor suppressor protein mutations (for example, those in Brca1).8, 9
Brca1 is a product of a breast and ovarian cancer susceptibility gene and participates in a variety of cellular processes, including DNA damage repair, cell cycle checkpoint response, chromatin remodeling and mitotic progression regulation.10 The involvement of Brca1 in centrosome regulation was recognized upon analysis of BRCA1−/− mice fibroblasts, which contain amplified centrosomes.11 Brca1 localizes to centrosomes during mitosis, and its ubiquitin ligase activity ensures normal centrosome number and function.9, 12 The direct target of Brca1 in the centrosome is γ-tubulin.8 Cells that express mutant γ-tubulin that cannot be ubiquitinated by Brca1 are characterized by centrosome amplification; a Brca1 mutant that lacks E3-ligase enzyme activity also loses its effect on centrosome nucleation inhibition.12, 13, 14
Chk2 is another centrosome localization tumor suppressor. In response to DNA double-stranded breaks, DNA damage sensor kinase ataxia telangiectasia mutated (ATM) phosphorylates Chk2 at Thr68 residue promoting Chk2 homodimerization and full activation.15 Phosphorylated Chk2 Thr68 reportedly localizes at centrosomes during mitosis;16 however, its role in mitotic regulation has not yet been established. Stolz et al.17 recently demonstrated that Chk2 is required for proper and timely spindle formation during normal mitosis; this is critical for accurate attachment of kinetochores to the spindle microtubules and subsequent proper chromosome segregation. Chk2 phosphorylates Ser988 of Brca1, which is important for the function of Brca1 in switching error-prone non-homologous end-joining (NHEJ) to error-free homologous recombination.18, 19, 20 Chk2 also phosphorylates Brca1 on Ser988 during normal mitotic progression in the absence of DNA damage. Wild-type Brca1, but not the Ser988 to alanine mutant, can rescue the mitosis defect induced by Chk2 deficiency, suggesting that phosphorylation of Brca1 at this site has an important role in mitotic spindle assembly and chromosome stability maintenance. Moreover, Chk2 deficiency increases CIN and leads to aneuploidy even in chromosomal-stable human colon carcinoma HCT116 cells.17 How Chk2 is activated in normal mitosis is still an unanswered question.
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is an essential component in the NHEJ pathway of double-stranded DNA break repair.21 In response to double-stranded breaks, DNA-PKcs is rapidly phosphorylated on its Thr2609 and Ser2056 clusters by ATM and by itself.22, 23 In addition to its established role in DNA repair, we observed that DNA-PKcs phosphorylated at both Thr2609 and Ser2056 localizes at the mitotic spindle apparatus on centrosomes and kinetochores. Consistent with this localization, DNA-PKcs is important for the stabilization of centrosome and of the spindle structure, as well as in the regulation of mitotic progression.24 DNA-PKcs regulates mitotic catastrophe in response to irradiation at least partially via the Chk2 pathway;25 however, whether Chk2 is a target of DNA-PKcs during normal mitotic progression and its biological significance in mitotic spindle assembly has not been clarified.
In the present study, we demonstrate that DNA-PKcs regulates Chk2 phosphorylation at the Thr68 site during mitosis and that DNA-PKcs coordinates the spindle assembly and kinetochore/microtubule attachment through the Chk2–Brca1 signaling pathway. We also show that the deficiency of DNA-PKcs leads to chromosomal instability.
These results demonstrate that DNA-PKcs mediates Chk2 phosphorylation to regulate microtubule nucleation and the spindle damage response.
Results
Chk2 is phosphorylated at Thr68 by DNA-PKcs during mitosis
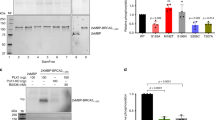
Chk2 is the key downstream effector of the ATM signaling pathway for cell cycle checkpoint regulation in the DNA damage response.26 In addition, Chk2 is activated during normal mitosis, as monitored by Thr68 phosphorylation, and is required for proper mitotic spindle assembly and chromosomal stability.27 We have reported that DNA-PKcs is also activated and has a critical role in mitosis progression.21 To test the hypothesis that DNA-PKcs is involved in Chk2 activation during mitosis, human colon cancer HCT116 cells and isogenic DNA-PKcs knockout cells (DNA-PKcs−/−) were mitotically synchronized or were treated with ionizing radiation. Western blot analyses revealed significant Chk2 Thr68 phosphorylation in HCT116 cells but little in DNA-PKcs−/− cells (Figure 1a). In contrast, ionizing radiation-induced Chk2 Thr68 phosphorylation (mediated by the ATM kinase) was observed in DNA-PKcs−/− cells at levels comparable to those in HCT116 cells. Similarly, siRNA-mediated depletion of DNA-PKcs from HeLa cells abolished the increase of Chk2 Thr68 phosphorylation in mitotically synchronized cells but not in irradiated cells (Figure 1b). Mitosis-dependent Chk2 phosphorylation was also clearly detected in the isogenic and NHEJ-defective Ligase4−/− cells (Supplementary Figure S1A), indicating that Chk2 phosphorylation in mitosis is independent of DSB repair regulation.
DNA-PKcs is required for Chk2 Thr68 phosphorylation during mitosis. (a) Human colon cancer HCT116 cells and derivative DNA-PKcs−/− cells were synchronized with nocodazole (50 ng/ml, 16 h) or were irradiated (4 Gy, 1 h). Whole-cell lysates were separated by electrophoresis and western blotted with the indicated antibodies. (b) HeLa cells transfected with a control siRNA or with an siRNA against DNA-PKcs were synchronized with nocodazole (50 ng/ml, 16 h) or were irradiated (4 Gy, 1 h). Whole-cell lysates were separated by electrophoresis and were western blotted with the indicated antibodies. Asynchronous (Asy), mitosis enriched (M), γ-ray irradiation (IR).
To further examine the role of the kinase activity of DNA-PKcs on Chk2 activation in mitosis, the parental HCT116 cells were synchronized via nocodazole treatment and then they were incubated for 2 h with DNA-PKcs kinase inhibitor Nu7441. As shown in Figure 2a, Nu7441 effectively abolished Chk2 Thr68 phosphorylation in response to mitosis synchrony but did not affect the levels of Thr68 phosphorylation induced by irradiation. The blocking of mitotic Chk2 phosphorylation was not due to alternation in mitosis synchrony as histone H3 Ser10 phosphorylation was equivalent in sham- and Nu7441-treated HeLa cells. Mitotic Chk2 phosphorylation was also elicited by treatment of cells with the microtubule stabilizer paclitaxel. Paclitaxel-mediated Chk2 Thr68 phosphorylation was abolished by treatment of cells with an siRNA targeted against the gene encoding DNA-PKcs or by Nu7441 treatment (Supplementary Figures S1B and C, respectively). In contrast, ATM kinase inhibitor Ku55933 had no effect on mitosis-induced Chk2 phosphorylation but did effectively block IR-induced Chk2 phosphorylation (Figure 2b). Taken together, these results demonstrate that DNA-PKcs, but not the ATM kinase, activates Chk2 during mitosis.
DNA-PKcs kinase inhibition attenuates mitotic induction of Chk2 phosphorylation. (a) Wild-type HCT116 cells were treated with nocodazole for 16 h, followed by a 2 h incubation with DMSO or 10 μM Nu7441 (Nu). (b) HeLa cells were subjected to nocodazole (50 ng/ml, 16 h), followed by a 2 h incubation with DMSO, Nu7441, Ku55933 or both Nu7441 and Ku55933 (left panel). HeLa cells were also irradiated (4 Gy, 1 h) with or without pretreatment with Nu7441 and Ku55933 (right panel).
DNA-PKcs is required for mitotic spindle assembly and chromosomal stability
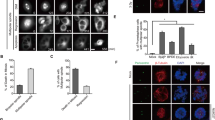
During the prometaphase to metaphase transition, kinetochores of sister chromatid pairs are captured by microtubules of opposite poles to allow chromosomes to align at the center of the spindle before the sister-chromatid separation at anaphase. Stolz et al.17 reported that Chk2 activation is necessary for proper spindle organization and chromosome segregation during normal mitosis and that Chk2 deficiency resulted in chromosomal numerical instability. Similarly, we observed that there was an increase of chromosomal numerical instability in DNA-PKcs−/− cells as compared with the parental HCT116 cells and Ligase4−/− cells (Figure 3a). To further explore the involvement of DNA-PKcs in mitotic spindle assembly, HCT116, DNA-PKcs−/− and Ligase4−/− cells were treated with nocodazole and then with proteasome inhibitor MG132 to stall mitosis progress at the metaphase to anaphase transition. Abnormal mitotic spindles were examined via immunofluorescence staining. Cells with misaligned chromosomes were scored (Figure 3b). DNA-PKcs−/− cells had a higher proportion of chromosomal misalignment during the metaphase to anaphase transition than did the parental HCT116 cells or the Ligase4−/− cells (Figure 3d).
Loss of DNA-PKcs increases mitosis defects and chromosomal instability. (a) Chromosome numbers from individual metaphase spreads of HCT116, DNA-PKcs−/− and Ligase4−/− cells (n⩾100). (b) HCT116, DNA-PKcs−/− and Ligase4−/− cells were treated with 50 ng/ml nocodazole for 16 h, and, after release into fresh medium, they were treated with 10 μM MG132 for 3 h. Cells were stained for α-tubulin (red), crest (green) and DNA (blue). (c) Exponentially growing HCT116, DNA-PKcs−/− and Ligase4−/− cells were subjected to the same immunofluorescent staining protocol. Arrowheads indicate lagging chromosome. (d) Percentage of mitotic cells with misaligned chromosomes from three independent analyses. (e) Percentages of mitotic cells showed lagging chromosome were counted from three independent analyses. **P<0.01.
Abnormal mitotic spindles were further examined via analysis of chromosome lagging in anaphase cells, an indication of chromosomal instability or errors during chromosomal segregation. Exponentially growing HCT116, DNA-PKcs−/− and Ligase4−/− cells were scored for chromosome lagging in anaphase (Figure 3c). Lack of DNA-PKcs, but not of Ligase4, caused a significant increase of lagging chromosomes in HCT116 cells (Figure 3e), which is consistent with the increase of aneuploidy found in DNA-PKcs−/− cells but not in Ligase4−/− cells (Figure 3a). Transient knockdown of levels of DNA-PKcs using an siRNA or treatment with DNA-PKcs inhibitor Nu7441 also promoted abnormal spindles and lagging chromosomes in HeLa cells (Supplementary Figure S2). These results further demonstrated the importance of DNA-PKcs and its kinase activity in mitotic spindle assembly and maintenance of chromosomal stability.
The Chk2–Brca1 pathway mediates DNA-PKcs signaling in spindle organization
To further test the function of DNA-PKcs-dependent Chk2 phosphorylation in spindle assembly and chromosomal stability, wild-type Chk2 and Chk2 mutants with an alanine (to prevent phosphorylation) or aspartic acid (to mimic the phosphorylated state) substitution at Thr68 residue (T68A and T68D, respectively) were expressed in DNA-PKcs−/− cells (Figure 4a). Overexpression of wild-type Chk2 or of the T68A mutant did not alter the frequencies of chromosomal misalignment and lagging in DNA-PKcs−/− cells as compared with control cells transfected with empty vector, whereas overexpression of the phosphomimetic Chk2 T68D mutant significantly decreased the levels of these abnormalities during mitosis (Figure 4b). T68D mutant Chk2 was able to restore proper chromosomal alignment and alleviate anaphase lagging in HCT116 cells, whereas vector alone had no effect. Chk2 is known to phosphorylate Brca1 at Ser988 upon DNA damage, which facilitates Brca1 function in DSB repair.18, 19, 20 The involvement of Chk2 signaling in mitosis and spindle assembly has also been linked to Brca1 Ser988 phosphorylation.17 To test whether Brca1 also functions in the same signaling pathway downstream from DNA-PKcs, wild-type Brca1 and non-phosphorylatable or phosphomimetic Brca1 mutants (S988A and S988E, respectively) were overexpressed in DNA-PKcs−/− cells (Figure 4c). As was observed in Chk2 overexpression experiments, the phosphomimetic Brca1 mutant, but not wild-type Brca1 nor the S988A mutant, was able to rescue mitotic defects in DNA-PKcs−/− cells (Figure 4d). Taken together, our analyses revealed that Chk2 and Brca1 are the downstream effectors of DNA-PKcs signaling in mitotic regulation.
Phosphomimetic Chk2 and BRCA1 mutants partially rescue chromosomal instability in DNA-PKcs-deficient cells. (a) DNA-PKcs−/− cells were transfected with constructs for the expression of flag-tagged Chk2 (wild-type, T68A, T68D). Expression of exogenous flag-tagged Chk2 and endogenous Chk2 was evaluated by western blot analysis with α-Chk2 antibody. (b) Aberrances in mitosis were analyzed in control and cells expressing flag-Chk2 in two independent experiments. (c) DNA-PKcs−/− cells were transfected with Brca1 constructs (wild-type, S988A, and S988E). Expression of wild-type and mutant Brca1 was evaluated by western blot. (d) Aberrances in mitosis were calculated from two independent experiments. *P<0.05; **P<0.01.
DNA-PKcs inactivation leads to dysfunction of microtubule dynamics
Improper regulation of microtubule dynamics is associated with aberrant mitotic spindle and chromosomal instability.28 The Chk2–Brca1 signaling axis has also been implicated in regulation of microtubule dynamics in response to spindle damage.29, 30 To further investigate the involvement of DNA-PKcs in microtubule regulation, HCT116 and DNA-PKcs−/− cells were treated with nocodazole in chilled medium to depolymerize microtubules. Microtubule nucleation and regrowth was initiated upon removal of nocodazole-containing medium and addition of prewarmed fresh medium. Microtubule nucleation status was inferred based on the diameter of asters, the radial arrays of microtubules that grow from centrosomes (Figure 5a). Our analyses revealed that microtubule growth was significantly enhanced in DNA-PKcs−/− relative to HCT116 cells (Figure 5b). To determine the role of Chk2 and DNA-PKcs in microtubule regulation, HeLa cells stably expressing wild-type or T68A or T68D mutant Chk2 were transfected with siRNA targeting the gene encoding DNA-PKcs. Cells were analyzed for microtubule regrowth as described above. Depletion of DNA-PKcs caused a significant outgrowth of microtubules in cells expressing wild-type Chk2, the T68A mutant and the vector control but not in cells expressing Chk2 T68D (Figure 5c and Supplementary Figure S3). These results indicate that the phosphomimetic Chk2 T68D mutant protein can compensate for the loss of DNA-PKcs and prevent dysregulation of microtubule dynamics in mitosis.
Phosphomimetic Chk2 alleviates dysregulation of microtubule nucleation in DNA-PKcs−/− cells. (a) HCT116 and DNA-PKcs−/− cells were treated with nocodazole to disrupt microtubules. Microtubule nucleation and regrowth was monitored at the indicated time points after nocodazole removal. (b) The length of the microtubule emanating from the centrosomes was measured (n⩾50). (c) HeLa cells expressing wild-type Chk2 or T68A or T68D mutant Chk2 were transfected with control or DNA-PKcs-targeted siRNA. Forty-eight hours after transfection, cells were subjected to microtubule nucleation analysis (n⩾50). *P<0.05; **P<0.01.
Microtubule formation in Brca1-deficient cancer cells is abnormal when cells are treated with an anti-microtubule agent such as paclitaxel.31, 32 The connection of DNA-PKcs and Chk2–Brca1 signaling in regulation of microtubule dynamics prompted us to speculate that DNA-PKcs modulates cellular sensitivity toward paclitaxel. To test the hypothesis, HCT116 and DNA-PKcs−/− cells were treated with paclitaxel and were subjected to a clonogenic survival assay. Our analysis revealed that DNA-PKcs deficiency in HCTT116 cells did confer paclitaxel resistance in clonogenic survival assay (Figure 6a). In addition, the colony sizes of HCT116 cells were obviously smaller than those of DNA-PKcs−/− cells (data not shown). Similarly, an MTT cell viability assay revealed that DNA-PKcs−/− cells were more resistant to paclitaxel than were the parental HCT116 cells (Figure 6b).
DNA-PKcs deficiency renders cells resistant to paclitaxel. (a) HCT116 and DNA-PKcs−/− cells were treated with indicated concentrations of paclitaxel and were analyzed for clonal survival ability. (b) HCT116 and DNA-PKcs−/− cells were treated with paclitaxel for 72 h, and viability was determined using an MTT assay. ***P<0.001; ****P<0.0001.
Discussion
We have previously reported that DNA-PKcs is autophosphorylated during normal cell cycle progression into mitosis and that activation of DNA-PKcs is necessary for microtubule organization at centrosomes and kinetochores, mitotic spindle organization and chromosomal segregation.24 In the current study, we report that DNA-PKcs is the upstream regulator of the Chk2–Brca1 signaling pathway, which was known to regulate mitotic spindle formation and maintenance of chromosomal stability. Our results demonstrate that mitotic induction of Chk2 phosphorylation at Thr68 is primarily mediated by DNA-PKcs; inhibition of DNA-PKcs via different approaches eliminated mitotic Chk2 phosphorylation. This is in contrast to ATM-dependent Chk2 phosphorylation under the DNA damage conditions.33 Chk2 activation has been linked to its downstream effector molecule Brca1 in both DNA damage response and normal mitosis progression, and Chk2-dependent Brca1 phosphorylation at Ser988 is critical for proper assembly of the mitotic spindle and for chromosomal stability.17 Our analyses revealed that overexpression of phosphomimetic mutants of Chk2 (T68D) or Brca1 (S988E) but not their wild-type counterparts compensated for the loss of DNA-PKcs and decreased chromosomal misalignment and lagging in cells that were deficient in DNA-PKcs (Figures 4 and 5). These results demonstrate that DNA-PKcs, Chk2 and Brca1 are functionally aligned in the pathway that regulates mitotic spindle assembly and guides accurate chromosomal segregation. Our data do not rule out the possibility that, in the absence of DNA-PKcs, a compromised NHEJ mechanism in DSB repair results in aberrant chromosomal structures that hinder chromosomal alignment and segregation in mitosis. However, we did not observe a significant increase in aberrant mitosis in NHEJ-deficient ligase4−/− cells, suggesting that a mechanism independent of both NHEJ and DSB repair but dependent on DNA-PKcs phosphorylation of Chk2 regulates mitosis.
The presence of mitotic DNA-PKcs phosphorylation at centrosomes and kinetochores implies that DNA-PKcs is involved in microtubule organization and could modulate k-MT attachment.24 Accurate chromosome segregation in mitosis relies on the sister kinetochores attached to microtubules emanating from opposite spindle poles. Inaccurate and aberrant k-MT attachments result in CIN, aneuploidy and full-blown carcinogenesis.34, 35 There are three types of k-MT attachment errors in prometaphase: (1) monotelic attachment, in which only one sister kinetochore attaches to microtubule emanating from one pole; (2) syntelic attachment, in which both sister kinetochores bind to microtubules from the same pole; and (3) merotelic attachment, in which a kinetochore is attached to microtubules from both poles. The merotelic attachment is the only attachment error that can escape from the surveillance of the spindle checkpoint, as no kinetochore is left unattached.36, 37 Therefore, cells carrying merotelic kinetochore attachment can progress into anaphase with lagging chromosomes; this is the most common route to gain or loss of a complete chromosome and results in aneuploidy.6, 35
Loss of Brca1 or Chk2 coincides with increased incidents of aneuploidy and merotelic kinetochore attachment.27 Similarly, there is an increase of misalignment and lagging chromosomes in the absence of DNA-PKcs, and overexpression of phosphomimetic Chk2 T68D or Brca1 S988E partially rescued these aberrances in DNA-PKcs-deficient cells. Thus, it is tempting to speculate that the DNA-PKcs-Chk2–Brca1 pathway triggers correction of merotelic k-MT attachments. Early studies revealed that Brca1 resides at the centrosome throughout all cell cycle phases and suppresses centrosome amplification, microtubule nucleation and aneuploidy.30, 38 Brca1 function in centrosome and microtubule regulation requires its dimerization with Brca1-associated ring domain protein 1 (Bard1) to form an active E3 ubiquitin ligase.8 A key target of Brca1-dependent ubiquitination is γ-tubulin, which is a component of the γ-tubulin ring complex that anchors microtubule nucleation and growth.39 The Brca1/Brad1 complex directs γ-tubulin monoubiquitination at lysines 48 and 344 in S/G2 phases, and failure of γ-tubulin monoubiquitination results in a marked amplification of centrosomes and an increase in microtubule nucleation.12, 14 These results thus recapitulate centrosome hyperactivation in Brca1-deficient cells and support a negative regulatory function of Brca1 on microtubule regulation. Such an inhibitory effect on microtubule also requires Chk2-mediated Brca1 phosphorylation at Ser988.29
Consistent with this model, microtubule regrowth analysis revealed that an enhancement of microtubule nucleation/regrowth occurs in DNA-PKcs-deficient cells and that such dysregulation can be corrected by expression of the phosphomimetic Chk2 T68D mutant (Figure 5 and Supplementary Figure S3). It is likely that, in the absence of DNA-PKcs, inadequate activation of the Chk2–Brca1 signaling pathway leads to excessive microtubule nucleation and aberrant microtubule–kinetochore attachment. It has been reported that loss of Brca1 in cancer cells confers resistance to paclitaxel treatment,40, 41 which causes microtubule stabilization and mitotic cell death.42 Similarly, loss of DNA-PKcs is also associated with paclitaxel resistance, thus further supporting the notion that DNA-PKcs regulates microtubule dynamics via the Chk2–Brca1 pathway.
The role of the Chk2–Brca1 pathway in k-MT attachments could be modulated or compensated by the Aurora kinases. Aurora-B kinase is known to have an important role in correcting aberrant microtubule attachment through phosphorylation/activation of the mitotic centromere-associated kinesin (microtubule depolymerizing enzyme MCAK), which depolymerizes microtubules.43, 44 Overexpression of MCAK suppresses lagging chromosomes and aneuploidy owing to the loss of Chk2.17 However, our result indicates that mitotic activation of DNA-PKcs is independent of Aurora-B regulation (data not shown), suggesting that the DNA-PKcs-Chk2–Brca1 axis acts parallel to the Aurora-B pathway to suppress aberrant microtubule attachment and facilitate chromosomal stability. It has also been reported that Aurora-A kinase phosphorylates Brca1 during mitosis and attenuates its E3 ubiquitin ligase activity.45 It is possible that DNA-PKcs and Aurora-A are required for a balanced Brca1 activity for optimal regulation of microtubule dynamics and k-MT attachments in mitosis.
It is conceivable that the defect in this DNA-PKcs-Chk2–Brca1 signaling pathway will result in chromosomal instability, which not only is a hallmark of cancer cells but also an early event and the driving force for tumorigenesis.46 The connection of Chk2 and Brca1 to cancer development has been well documented, as they have been recognized as multiorgan cancer susceptibility genes.47, 48 In addition, a growing body of evidence has implicated a role of DNA-PKcs in tumorigenesis.49 For example, progressive loss of DNA-PKcs expression has been correlated to the advancement in ovarian cancer development.50 Loss of DNA-PKcs expression has also been found in gastric tumors and correlated to a frameshift mutation of the poly(A)10 tract, as well as microsatellite instability, tumor progression and poor patient survival.51, 52 In addition, reduction in DNA-PKcs activity in peripheral blood lymphocytes has been associated with an increase in chromosomal instability and cancer risk.53, 54 Consistent with these analyses, DNA-PKcs kinase inhibition with small-molecule inhibitor not only attenuated mitotic Chk2 phosphorylation (Figure 1) but also impaired chromosomal alignment and prolonged mitosis progression.24 The current study thus provides a molecular mechanism that DNA-PKcs-dependent Chk2–Brca1 signaling pathway is critical for proper chromosomal segregation during mitosis and contributes as a barrier against chromosomal instability and tumorigenesis.
In summary, our study suggests that DNA-PKcs is the upstream activator of Chk2 and Brca1. The DNA-PKcs-Chk2–Brca1 pathway has an important role in the maintenance of chromosome stability through the regulation of centrosome homeostasis and proper microtubule attachment to the kinetochore. Our results also highlight the role of DNA-PKcs in the control of microtubule dynamics and the response to microtubule damage.
Materials and methods
Cell lines and treatment
Human cervical cancer HeLa cells, human colorectal carcinoma HCT116 cells and derivative cells55 were maintained in α-minimum essential medium supplemented with 10% fetal bovine serum and penicillin/streptomycin in a humidified incubator at 37 °C with 5% CO2. For mitosis synchrony, cells were treated with 50 ng/ml nocodazole (Sigma, St Louis, MO, USA) for 16 h; in certain experiments, cells were treated for an additional 2 h with kinase inhibitors before harvest. Cells were treated with ionizing radiation using a Mark-II Cesium-137 irradiator (J L Shepherd and Associates) with or without a 30-min pretreatment with kinase inhibitors. Cells were harvested at 1 h after irradiation. Cell transfection with siRNA oligonucleotides or constructs for expression of Chk2 or Brca118 was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Small inhibitory RNA (siRNA) oligonucleotides against DNA-PKcs were previously described.24
Immunoblotting and antibodies
Whole-cell lysate preparation and western blotting were performed as previously described.22, 24 For immunofluorescent staining, cells were grown on poly-D-lysine-coated culture slides (BD Pharmingen, San Diego, CA, USA), washed in phosphate-buffered saline (PBS), fixed in PBS containing 4% paraformaldehyde, permeabilized in 0.5% Triton X-100 and blocked in PBS containing 5% bovine serum albumin. The cells were incubated with indicated primary antibodies for 2 h at room temperature, washed with PBS and incubated with Alexa-568- and Alexa-488-conjugated secondary antibodies for 1 h (Invitrogen). Cells were then washed with PBS and mounted in Vectashield mounting medium with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, USA). Images were acquired from a Zeiss AxioImager M2 microscope system equipped with a Plan-Apochromat 63 × /NA 1.40 objective, an AxioCam MRm CCD camera and AxioVision software (Carl Zeiss, Oberkochen, Germany). Anti-Chk2 total (Cell Signaling, Beverly, MA, USA), anti-phosphorylated Chk2 at Thr68 (Cell Signaling), anti-phospho-histone H3 (EMD Millipore, Billerica, MA, USA), anti-β-actin (Sigma) and anti-Brca1 total (Santa Cruz, Dallas, TX, USA) antibodies were purchased from the indicated vendors. Antibodies against total and phosphorylated forms of DNA-PKcs were described previously.22
Microtubule nucleation assay
Cells were treated with cold medium containing 10 μg/ml nocodazole for 40 min, washed with PBS and added to prewarmed fresh medium without nocodazole to allow microtubule regrowth. Cells were fixed at the indicated time and subjected to immunofluorescent staining as described above.
Clonogenic survival and MTT cell proliferation assays
Exponentially growing cells were trypsinized and counted. HCT116 cells were diluted serially to appropriate concentrations, plated into 60-mm dishes in triplicate and treated with paclitaxel (0.5, 1, 2 and 6 nM). Cells were fixed at 10–14 days and were stained with 4% formaldehyde in PBS containing 0.05% crystal violet. Colonies containing >50 cells were counted. For cell proliferation assays, 1 × 104 cells were seeded per well in a 96-well plate. Cells were cultured with paclitaxel for 72 h, and then were analyzed for viability using the previously described MTT assay.56
References
Bakhoum SF, Compton DA . Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 2012; 122: 1138–1143.
Thompson SL, Bakhoum SF, Compton DA . Mechanisms of chromosomal instability. Curr Biol 2010; 20: R285–R295.
Qi W, Yu H . The spindle checkpoint and chromosomal stability. Genome Dyn 2006; 1: 116–130.
Cimini D, Moree B, Canman JC, Salmon ED . Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 2003; 116: 4213–4225.
Bakhoum SF, Genovese G, Compton DA . Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol 2009; 19: 1937–1942.
Ganem NJ, Godinho SA, Pellman D . A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460: 278–282.
Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL . Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA 1998; 95: 2950–2955.
Parvin JD . The BRCA1-dependent ubiquitin ligase, gamma-tubulin, and centrosomes. Environ Mol Mutagen 2009; 50: 649–653.
Kais Z, Parvin JD . Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther 2008; 7: 1540–1543.
Deng CX . BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 2006; 34: 1416–1426.
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266: 66–71.
Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP et al. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol 2004; 24: 8457–8466.
Sankaran S, Starita LM, Simons AM, Parvin JD . Identification of domains of BRCA1 critical for the ubiquitin-dependent inhibition of centrosome function. Cancer Res 2006; 66: 4100–4107.
Sankaran S, Starita LM, Groen AC, Ko MJ, Parvin JD . Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol Cell Biol 2005; 25: 8656–8668.
Ahn J, Urist M, Prives C . The Chk2 protein kinase. DNA Repair (Amst) 2004; 3: 1039–1047.
Tsvetkov L, Xu X, Li J, Stern DF . Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J Biol Chem 2003; 278: 8468–8475.
Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol 2010; 12: 492–499.
Wang HC, Chou WC, Shieh SY, Shen CY . Ataxia telangiectasia mutated and checkpoint kinase 2 regulate BRCA1 to promote the fidelity of DNA end-joining. Cancer Res 2006; 66: 1391–1400.
Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol 2004; 24: 708–718.
Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res 2006; 66: 1401–1408.
Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 2002; 16: 2333–2338.
Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 2005; 280: 14709–14715.
Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H et al. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 2007; 282: 6582–6587.
Lee KJ, Lin YF, Chou HY, Yajima H, Fattah KR, Lee SC et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem 2011; 286: 12796–12802.
Shang ZF, Huang B, Xu QZ, Zhang SM, Fan R, Liu XD et al. Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 Phosphorylation in response to DNA damage. Cancer Res 2010; 70: 3657–3666.
Smith J, Tho LM, Xu N, Gillespie DA . The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 2010; 108: 73–112.
Stolz A, Ertych N, Bastians H . Loss of the tumour-suppressor genes CHK2 and BRCA1 results in chromosomal instability. Biochem Soc Trans 2010; 38: 1704–1708.
Rao CV, Yamada HY, Yao Y, Dai W . Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis 2009; 30: 1469–1474.
Chabalier-Taste C, Racca C, Dozier C, Larminat F . BRCA1 is regulated by Chk2 in response to spindle damage. Biochim Biophys Acta 2008; 1783: 2223–2233.
Hsu LC, White RL . BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci USA 1998; 95: 12983–12988.
Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 2003; 63: 6221–6228.
Tassone P, Tagliaferri P, Perricelli A, Blotta S, Quaresima B, Martelli ML et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer 2003; 88: 1285–1291.
Shiloh Y, Ziv Y . The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14: 197–210.
Ruchaud S, Carmena M, Earnshaw WC . Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 2007; 8: 798–812.
Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D . Merotelic kinetochore attachment: causes and effects. Trends Cell Biol 2011; 21: 374–381.
O’Connell CB, Loncarek J, Hergert P, Kourtidis A, Conklin DS, Khodjakov A . The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol 2008; 183: 29–36.
Cimini D . Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta 2008; 1786: 32–40.
Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell 1999; 3: 389–395.
Luders J, Stearns T . Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 2007; 8: 161–167.
Chabalier C, Lamare C, Racca C, Privat M, Valette A, Larminat F . BRCA1 downregulation leads to premature inactivation of spindle checkpoint and confers paclitaxel resistance. Cell Cycle 2006; 5: 1001–1007.
Zhou C, Smith JL, Liu J . Role of BRCA1 in cellular resistance to paclitaxel and ionizing radiation in an ovarian cancer cell line carrying a defective BRCA1. Oncogene 2003; 22: 2396–2404.
Jordan MA, Wendell K, Gardiner S, Derry WB, Copp H, Wilson L . Mitotic block induced in HeLa cells by low concentrations of paclitaxel (Taxol) results in abnormal mitotic exit and apoptotic cell death. Cancer Res 1996; 56: 816–825.
Lampson MA, Cheeseman IM . Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 2011; 21: 133–140.
Kelly AE, Funabiki H . Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 2009; 21: 51–58.
Sankaran S, Crone DE, Palazzo RE, Parvin JD . Aurora-A kinase regulates breast cancer associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res 2007; 67: 11186–11194.
Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih IeM, Vogelstein B et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci USA 2002; 99: 16226–16231.
Antoni L, Sodha N, Collins I, Garrett MD . CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat Rev Cancer 2007; 7: 925–936.
Narod SA, Foulkes WD . BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 2004; 4: 665–676.
Hsu FM, Zhang S, Chen BP . Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res 2012; 1: 22–34.
Shao SL, Cai Y, Wang QH, Yan LJ, Zhao XY, Wang LX . Expression of GLUT-1, p63 and DNA-Pkcs in serous ovarian tumors and their significance. Zhonghua Zhong Liu Za Zhi 2007; 29: 697–700.
Lee HS, Choe G, Park KU, Park do J, Yang HK, Lee BL et al. Altered expression of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) during gastric carcinogenesis and its clinical implications on gastric cancer. Int J Oncol 2007; 31: 859–866.
Lee HS, Yang HK, Kim WH, Choe G . Loss of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) expression in gastric cancers. Cancer Res Treat 2005; 37: 98–102.
Someya M, Sakata K, Matsumoto Y, Yamamoto H, Monobe M, Ikeda H et al. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis 2006; 27: 117–122.
Someya M, Sakata KI, Matsumoto Y, Kamdar RP, Kai M, Toyota M et al. The association of DNA-dependent protein kinase activity of peripheral blood lymphocytes with prognosis of cancer. Br J Cancer 2011; 104: 1724–1729.
Ruis BL, Fattah KR, Hendrickson EA . DNA-PKcs regulates proliferation, telomere length and genomic stability in human somatic cells. Mol Cell Biol 2008; 28: 6182–6195.
van Meerloo J, Kaspers GJ, Cloos J . Cell sensitivity assays: the MTT assay. Methods Mol Biol 2011; 731: 237–245.
Acknowledgements
This work was supported by the National Institutes of Health (CA166677) and the Cancer Prevention Research Institute of Texas (RP110465-P1). We thank Dr Eric Hendrickson for providing the human HCT116 and derivative cell lines.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Shang, Z., Yu, L., Lin, YF. et al. DNA-PKcs activates the Chk2–Brca1 pathway during mitosis to ensure chromosomal stability. Oncogenesis 3, e85 (2014). https://doi.org/10.1038/oncsis.2013.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2013.49
Keywords
This article is cited by
-
CHK2 activation contributes to the development of oxaliplatin resistance in colorectal cancer
British Journal of Cancer (2022)
-
BRCA1 mutations in high-grade serous ovarian cancer are associated with proteomic changes in DNA repair, splicing, transcription regulation and signaling
Scientific Reports (2022)
-
CtBP1/2 differentially regulate genomic stability and DNA repair pathway in high-grade serous ovarian cancer cell
Oncogenesis (2021)
-
Assessment of DNA-PKcs kinase activity by quantum dot–based microarray
Scientific Reports (2018)
-
PIG3 promotes NSCLC cell mitotic progression and is associated with poor prognosis of NSCLC patients
Journal of Experimental & Clinical Cancer Research (2017)