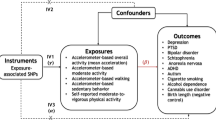
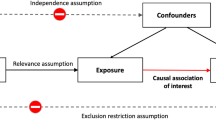
Abstract
Background/objectives
Physical activity (PA) protects against a wide range of diseases. Habitual PA appears to be heritable, motivating the search for specific genetic variants that may inform efforts to promote PA and target the best type of PA for each individual.
Subjects/methods
We used data from the UK Biobank to perform the largest genome-wide association study of PA to date, using three measures based on self-report (nmax = 377,234) and two measures based on wrist-worn accelerometry data (nmax = 91,084). We examined genetic correlations of PA with other traits and diseases, as well as tissue-specific gene expression patterns. With data from the Atherosclerosis Risk in Communities (ARIC; n = 8,556) study, we performed a meta-analysis of our top hits for moderate-to-vigorous PA (MVPA).
Results
We identified ten loci across all PA measures that were significant in both a basic and a fully adjusted model (p < 5 × 10−9). Upon meta-analysis of the nine top hits for MVPA with results from ARIC, eight were genome-wide significant. Interestingly, among these, the rs429358 variant in the APOE gene was the most strongly associated with MVPA, whereby the allele associated with higher Alzheimer’s risk was associated with greater MVPA. However, we were not able to rule out possible selection bias underlying this result. Variants in CADM2, a gene previously implicated in obesity, risk-taking behavior and other traits, were found to be associated with habitual PA. We also identified three loci consistently associated (p < 5 × 10−5) with PA across both self-report and accelerometry, including CADM2. We found genetic correlations of PA with educational attainment, chronotype, psychiatric traits, and obesity-related traits. Tissue enrichment analyses implicate the brain and pituitary gland as locations where PA-associated loci may exert their actions.
Conclusions
These results provide new insight into the genetic basis of habitual PA, and the genetic links connecting PA with other traits and diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fiuza-Luces C, Garatachea N, Berger NA, Lucia A. Exercise is the real polypill. Physiol. 2013;28:330–58.
US Surgeon General. Physical Activity and Health: A Report of the Surgeon General. S/N 017-023-00196-5. 1996.
Blair SN. Physical inactivity: the biggest public health problem of the 21st century. Br J Sports Med. 2009;43:1–2.
Kaplan GA, Strawbridge WJ, Cohen RD, Hungerford LR. Natural history of leisure-time physical activity and its correlates: associations with mortality from all causes and cardiovascular disease over 28 years. Am J Epidemiol. 1996;144:793–7.
Bauman AE, Reis RS, Sallis JF, Wells JC, Loos RJF, Martin BW. Correlates of physical activity: Why are some people physically active and others not? Lancet. 2012;380:258–71.
Trost SG, Owen N, Bauman AE, Sallis JF, Brown W. Correlates of adults’ participation in physical activity: review and update. Med Sci Sport Exerc. 2002;34:1996–2001.
den Hoed M, Brage S, Zhao JH, Westgate K, Nessa A, Ekelund U, et al. Heritability of objectively assessed daily physical activity and sedentary behavior. Am J Clin Nutr. 2013;98:1317–25.
Gielen M, Westerterp-Plantenga MS, Bouwman FG, Joosen AMCP, Vlietinck R, Derom C, et al. Heritability and genetic etiology of habitual physical activity: a twin study with objective measures. Genes Nutr. 2014;9:415.
Stubbe JH, Boomsma DI, Vink JM, Cornes BK, Martin NG, Skytthe A, et al. Genetic influences on exercise participation in 37.051 twin pairs from seven countries. PLoS ONE. 2006;1:e22.
Joosen AMCP, Gielen M, Vlietinck R, Westerterp KR. Genetic analysis of physical activity in twins. Am J Clin Nutr. 2005;82:1253–9.
Pérusse L, Tremblay A, Leblanc C, Bouchard C. Genetic and environmental influences on level of habitual physical activity and exercise participation. Am J Epidemiol. 1989;129:1012–22.
Lauderdale DS, Fabsitz R, Meyer JM, Sholinsky P, Ramakrishnan V, Goldberg J. Familial determinants of moderate and intense physical activity: a twin study. Med Sci Sports Exerc. 1997;29:1062–8.
Kaprio J, Koskenvuo M, Sarna S. Cigarette smoking, use of alcohol, and leisure-time physical activity among same-sexed adult male twins. Prog Clin Biol Res. 1981;69(Pt C):37–46.
Thompson PD, Tsongalis GJ, Ordovas JM, Seip RL, Bilbie C, Miles M, et al. Angiotensin-converting enzyme genotype and adherence to aerobic exercise training. Prev Cardiol. 2006;9:21–4.
Herring MP, Sailors MH, Bray MS. Genetic factors in exercise adoption, adherence and obesity. Obes Rev Engl. 2014;15:29–39.
Wilkinson AV, Gabriel KP, Wang J, Bondy ML, Dong Q, Wu X, et al. Sensation-seeking genes and physical activity in youth. Genes, Brain Behav. 2013;12:181–8.
Caldwell Hooper AE, Bryan AD, Hagger MS. What keeps a body moving? The brain-derived neurotrophic factor val66met polymorphism and intrinsic motivation to exercise in humans. J Behav Med. 2014;37:1180–92.
Lightfoot JT. Current understanding of the genetic basis for physical activity. J Nutr. 2011;141:526–30.
De Moor MHM, Liu Y-J, Boomsma DI, Li J, Hamilton JJ, Hottenga J-J, et al. Genome-wide association study of exercise behavior in Dutch and American adults. Med Sci Sports Exerc. 2009;41:1887–95.
Kim J, Min H, Oh S, Kim Y, Lee AH, Park T. Joint identification of genetic variants for physical activity in Korean population. Int J Mol Sci. 2014;15:12407–21.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med Public Libr Sci. 2015;12:e1001779.
The Aric Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702.
Craig CL, Marshall AL, Sjöström M, Bauman AE, Booth ML, Ainsworth BE, et al. International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc. 2003;35:1381–95.
IPAQ Research Committee. Guidelines for Data Processing and Analysis of the International Physical Activity Questionnaire (IPAQ) - Short Form. J Am Diet Assoc. 2002
Ekelund U, Sepp H, Brage S, Becker W, Jakes R, Hennings M, et al. Criterion-related validity of the last 7-day, short form of the International Physical Activity Questionnaire in Swedish adults. Public Health Nutr. 2006;9:258–65.
American Heart Association. American Heart Association Recommendations for Physical Activity in Adults. 2016. http://www.heart.org/HEARTORG/HealthyLiving/PhysicalActivity/FitnessBasics/American-Heart-Association-Recommendations-for-Physical-Activity-in-Adults_UCM_307976_Article.jsp#.WqF2eOjwbIV. Accessed 20 Aug 2003.
Doherty A, Jackson D, Hammerla N, Plötz T, Olivier P, Granat MH, et al. Large scale population assessment of physical activity using wrist worn accelerometers: The UK Biobank Study. Buchowski M, editor. PLoS ONE. 2017;12:e0169649.
Hildebrand M, Van Hees VT, Hansen BH, Ekelund U. Age group comparability of raw accelerometer output from wrist-and hip-worn monitors. Med Sci Sports Exerc. 2014;46:1816–24.
Baecke JA, Burema J, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr. 1982;36:936–42.
Folsom AR, Arnett DK, Hutchinson RG, Liao F, Clegg LX, Cooper LS. Physical activity and incidence of coronary heart disease in middle-aged women and men. Med Sci Sport Exerc. 1997;29:901–9.
Richardson MT, Ainsworth BE, Wu HC, Jacobs DRJ, Leon AS. Ability of the Atherosclerosis Risk in Communities (ARIC)/Baecke Questionnaire to assess leisure-time physical activity. Int J Epidemiol Engl. 1995;24:685–93.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv. 2017
O’Connell J, Sharp K, Shrine N, Wain L, Hall I, Tobin M, et al. Haplotype estimation for biobank-scale data sets. Nat Genet. 2016;48:817–20.
Consortium GP, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83.
UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90.
Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ, et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98:456–72.
Howie B, Marchini J, Stephens M, Chakravarti A. Genotype Imputation with Thousands of Genomes. G3 GenesGenomesGenetics. 2011;1:457–70.
Townsend P. Deprivation. J Soc Policy. 1987;16:125–46. 2009/01/01. Cambridge University Press
Smith GD, Whitley E, Dorling D, Gunnell D. Area based measures of social and economic circumstances: cause specific mortality patterns depend on the choice of index. J Epidemiol Community Health. 2001;55:149–150.
Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90.
Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed model association for biobank-scale data sets. bioRxiv. Cold Spring Harbor Laboratory; 2017
Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–7.
Klarin D, Emdin CA, Natarajan P, Conrad MF, Kathiresan S Genetic Analysis of Venous Thromboembolism in UK Biobank Identifies the ZFPM2 Locus and Implicates Obesity as a Causal Risk FactorCLINICAL PERSPECTIVE. Circ Cardiovasc Genet. 2017;10:e001643. http://circgenetics.ahajournals.org/content/10/2/e001643.long https://doi.org/10.1161/CIRCGENETICS.116.001643
Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. 2016;24:1202–5.
Yaghootkar H, Bancks MP, Jones SE, McDaid A, Beaumont R, Donnelly L, et al. Quantifying the extent to which index event biases influence large genetic association studies. Hum Mol Genet Engl. 2017;26:1018–30.
Saunders AM, Strittmatter WJ, Schmechel D, St. George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele ϵ4 with late-onset familial and sporadic alzheimer’s disease. Neurology. 1993;43;1467–72.
Lahoz C, Schaefer EJ, Cupples LA, Wilson PW, Levy D, Osgood D, et al. Apolipoprotein E genotype and cardiovascular disease in the Framingham Heart Study. Atheroscler Irel. 2001;154:529–37.
Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol. 2002;155:487–95.
Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. mediation: R Package for Causal Mediation Analysis. J Stat Softw. 2014;59:1–38.
Team RDC. R: A language and environment for statistical computing. 2011.
Chyou PH A simple and robust way of concluding meta-analysis results using reported P values, standardized effect sizes, or other statistics. Clin. Med. Res. 2012;10:219–23.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D FUMA: Functional mapping and annotation of genetic associations. bioRxiv. 2017
Consortium TGte. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–60.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. Tang H, editor. PLOS Comput Biol. 2015;11:e1004219.
Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Consortium SWG of the PG. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Elliott L, Sharp K, Alfaro-Almagro F, Douaud G, Miller K, Marchini J, et al. The genetic basis of human brain structure and function: 1,262 genome-wide associations found from 3,144 GWAS of multimodal brain imaging phenotypes from 9,707 UK Biobank participants. bioRxiv. Cold Spring Harbor Laboratory; 2017 https://www.biorxiv.org/content/early/2017/08/25/178806 https://doi.org/10.1101/178806
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(1546–1718 (Electronic)):937–48.
Boutwell B, Hinds D, Tielbeek J, Ong KK, Day FR, Perry JRB. Replication and characterization of CADM2 and MSRA genes on human behavior. Heliyon Engl. 2017;3:e00349.
Day FR, Helgason H, Chasman DI, Rose LM, Loh P-R, Scott RA, et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet. 2016;48:617–23.
Strawbridge RJ, Ward J, Cullen B, Tunbridge EM, Hartz S, Bierut L, et al. Genome-wide analysis of risk-taking behaviour and cross-disorder genetic correlations in 116,255 individuals from the UK Biobank cohort. bioRxiv. 2018;8:39. https://www.nature.com/articles/s41398-017-0079-1
Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N = 112 117). Mol Psychiatry Engl. 2017;22:1376–84.
Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC, et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene [Internet]. Mol Psychiatry. 2016;21:189–97.
Yan X, Wang Z, Schmidt V, Gauert A, Willnow TE, Heinig M, et al. Cadm2 regulates body weight and energy homeostasis in mice. Mol Metab. 2017;8:180–8.
Pericak-Vance MA, Bebout JL, Gaskell PC Jr., Yamaoka LH, Hung WY, Alberts MJ, et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. AmJHumGenet. 1991;48:1034–50.
Thompson PD, Tsongalis GJ, Seip RL, Bilbie C, Miles M, Zoeller R, et al. Apolipoprotein e genotype and changes in serum lipids and maximal oxygen uptake with exercise training. Metabolism. 2004;53:193–202.
Dudbridge F, Fletcher O. Gene-environment dependence creates spurious gene-environment interaction. Am J Hum Genet. 2014;95:301–7.
Lee H, Ash GI, Angelopoulos TJ, Gordon PM, Moyna NM, Visich PS, et al. Obesity-related genetic variants and their associations with physical activity. Sport Med Open. 2015;1:34.
Richmond RC, Davey Smith G, Ness AR, den Hoed M, McMahon G, Timpson NJ. Assessing causality in the association between child adiposity and physical activity levels: A Mendelian Randomization Analysis. PLoS Med. 2014;11:e1001618.
Day FR, Loh P-R, Scott RA, Ong KK, Perry JR. A Robust Example of Collider Bias in a Genetic Association Study. Am J Hum Genet. 2016;98:392–3.
Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96:329–39.
Vink JM, Boomsma DI, Medland SE, de Moor MHM, Stubbe JH, Cornes BK, et al. Variance components models for physical activity with age as modifier: a comparative twin study in seven countries. Twin Res Hum Genet. 2011;14:25–34.
Shungin D, Winkler T, Croteau-Chonka D, Ferreira T, Mägi R, Lakka T, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96.
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E, et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet. 2017;49:1107–12.
Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12:e1006125.
Acknowledgements
This research was conducted using the UK Biobank Resource under Application Number 15678. We thank the participants and organizers of the UK Biobank. We also thank the participants and organizers of the ARIC study. Data from ARIC was obtained from dbGaP through accession number phs000280.v2.p1. The authors would like to acknowledge support from the National Institute of Diabetes and Digestive and Kidney Diseases grant (K01DK095032), the National Institute on Aging (AG019610), the State of Arizona and Arizona Department of Health Services (ADHS), and the McKnight Brain Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Atherosclerosis Risk in Communities: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Funding for GENEVA was provided by National Human Genome Research Institute grant U01HG004402 (E. Boerwinkle). The authors thank the staff and participants of the ARIC study for their important contributions. LDHUB Acknowledgements: We gratefully acknowledge all the studies and databases that made GWAS summary data available: ADIPOGen (Adiponectin genetics consortium), C4D (Coronary Artery Disease Genetics Consortium), CARDIoGRAM (Coronary ARtery DIsease Genome wide Replication and Meta-analysis), CKDGen (Chronic Kidney Disease Genetics consortium), dbGAP (database of Genotypes and Phenotypes), DIAGRAM (DIAbetes Genetics Replication And Meta-analysis), ENIGMA (Enhancing Neuro Imaging Genetics through Meta Analysis), EAGLE (EArly Genetics & Lifecourse Epidemiology Eczema Consortium, excluding 23andMe), EGG (Early Growth Genetics Consortium), GABRIEL (A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community), GCAN (Genetic Consortium for Anorexia Nervosa), GEFOS (GEnetic Factors for OSteoporosis Consortium), GIANT (Genetic Investigation of ANthropometric Traits), GIS (Genetics of Iron Status consortium), GLGC (Global Lipids Genetics Consortium), GPC (Genetics of Personality Consortium), GUGC (Global Urate and Gout consortium), HaemGen (haemotological and platelet traits genetics consortium), HRgene (Heart Rate consortium), IIBDGC (International Inflammatory Bowel Disease Genetics Consortium), ILCCO (International Lung Cancer Consortium), IMSGC (International Multiple Sclerosis Genetic Consortium), MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium), MESA (Multi-Ethnic Study of Atherosclerosis), PGC (Psychiatric Genomics Consortium), Project MinE consortium, ReproGen (Reproductive Genetics Consortium), SSGAC (Social Science Genetics Association Consortium) and TAG (Tobacco and Genetics Consortium), TRICL (Transdisciplinary Research in Cancer of the Lung consortium), UK Biobank. We gratefully acknowledge the contributions of Alkes Price (the systemic lupus erythematosus GWAS and primary biliary cirrhosis GWAS) and Johannes Kettunen (lipids metabolites GWAS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Klimentidis, Y.C., Raichlen, D.A., Bea, J. et al. Genome-wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes 42, 1161–1176 (2018). https://doi.org/10.1038/s41366-018-0120-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-018-0120-3
This article is cited by
-
Potential causal associations between leisure sedentary behaviors, physical activity, sleep traits, and myopia: a Mendelian randomization study
BMC Ophthalmology (2024)
-
Clinically relevant plasma proteome for adiposity depots: evidence from systematic mendelian randomization and colocalization analyses
Cardiovascular Diabetology (2024)
-
Causal links between sedentary behavior, physical activity, and psychiatric disorders: a Mendelian randomization study
Annals of General Psychiatry (2024)
-
Association between modifiable lifestyle factors and telomere length: a univariable and multivariable Mendelian randomization study
Journal of Translational Medicine (2024)
-
Investigation of genetic determinants of cognitive change in later life
Translational Psychiatry (2024)