Abstract
The accurate and timely diagnosis of disease is a prerequisite for efficient therapeutic intervention and epidemiological surveillance. Diagnostics based on the detection of nucleic acids are among the most sensitive and specific, yet most such assays require costly equipment and trained personnel. Recent developments in diagnostic technologies, in particular those leveraging clustered regularly interspaced short palindromic repeats (CRISPR), aim to enable accurate testing at home, at the point of care and in the field. In this Review, we provide a rundown of the rapidly expanding toolbox for CRISPR-based diagnostics, in particular the various assays, preamplification strategies and readouts, and highlight their main applications in the sensing of a wide range of molecular targets relevant to human health.
Similar content being viewed by others
Main
The fast and accurate diagnosis of a disease is central to effective treatment and to the prevention of long-term sequelae1. Nucleic-acid-based biomarkers associated with disease are essential for diagnostics because DNA and RNA can be amplified from trace amounts, which enables their highly specific detection via the pairing of complementary nucleotides. In fact, nucleic-acid-based diagnostics have become the gold standard for various acute and chronic conditions, especially those caused by infectious diseases2. During infectious-disease outbreaks, as most recently experienced with the coronavirus disease 2019 (COVID-19) pandemic, fast and precise nucleic-acid-based testing is vital for effective disease control3,4. The detection of nucleic acid biomarkers is also critical for agriculture and food safety, for environmental monitoring and in the sensing of biological warfare agents.
Nucleic acid-based diagnostics relying on the quantitative polymerase chain reaction (qPCR) or on sequencing have been widely adopted, and are frequently used in clinical laboratories. The versatility, robustness and sensitivity of PCR have made this technology the most commonly used for the detection of DNA and RNA biomarkers. In fact, PCR is the gold-standard technique for most nucleic-acid-based diagnostics. However, the costs of reagents for PCR are high, and the technique requires sophisticated laboratory equipment and trained personnel5. Although isothermal nucleic acid amplification circumvents the need for thermal cyclers, non-specific amplification can result in lower detection specificity6. The specificity can be improved through additional readouts, in particular by fluorescent probes7, oligo strand-displacement probes8 or molecular beacons9. However, there is a need for technologies that combine the ease of use and cost efficiency of isothermal amplification with the diagnostic accuracy of PCR. Ideally, such next-generation diagnostics should also have single-nucleotide specificity, which is integral to the detection of mutations conferring resistance against antibiotics10 or antiviral drugs11.
Clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostics have the potential to fulfil these unmet needs. CRISPR systems are a fundamental part of a microbial adaptive immune system12 that recognizes foreign nucleic acids on the basis of their sequence to subsequently eliminate them by means of endonuclease activity associated with the CRISPR-associated (Cas) enzyme. Although there are diverse CRISPR–Cas systems among the different species of archaea and bacteria13, these systems are connected by their dependence on CRISPR RNA (crRNA), which guides Cas proteins to recognize and cleave nucleic acid targets. The crRNA can be programmed towards a specific DNA or RNA region of interest through hybridization to a complementary sequence, which in some systems is restricted to the proximity of a protospacer adjacent motif (PAM) or protospacer flanking sequence14. So far, CRISPR–Cas systems have been repurposed for a variety of applications, including the targeted editing of genomes15, epigenomes16 and transcriptomes17, the bioimaging of nucleic acids18, the recording of cellular events19 and the detection of nucleic acids. Overall, the fast-evolving area of CRISPR-based diagnostics builds on the specificity, programmability and ease of use of CRISPR technology, and aims to create nucleic-acid-based point-of-care (POC) diagnostic tests for use in routine clinical care.
In this Review, we outline how the properties of different Cas enzymes have been leveraged for diagnostic assays, discuss different preamplification strategies (which today form part of most CRISPR-based diagnostic assays), examine approaches for quantification and multiplexing, highlight technologies that combine CRISPR-based target enrichment with sequencing, and review the optimization of CRISPR-based diagnostics for POC applications, with a focus on assay readouts and sample preparation. We also provide an overview of the emerging biomedical applications of the technology, and discuss open challenges and opportunities.
CRISPR-based methods for disease diagnosis
Since their initial discovery, the number of different CRISPR–Cas systems has expanded rapidly13,20,21. Currently, CRISPR–Cas systems can be divided, according to evolutionary relationships, into two classes, six types and several subtypes20. The classes of CRISPR–Cas system are defined by the nature of the ribonucleoprotein effector complex: class 1 systems are characterized by a complex of multiple effector proteins, and class 2 systems encompass a single crRNA-binding protein. The design of crRNAs for the different effector proteins used in CRISPR diagnostics follow the same principles as those of other CRISPR applications, and are summarized in Table 1. Among the diverse CRISPR systems, class 2 systems have primarily been applied for diagnostics, as these systems are simpler to reconstitute. They include enzymes with collateral activity, which serves as the backbone of many CRISPR-based diagnostic assays (Table 2). Class 1 systems (such as the type III effector nuclease Csm6 or Cas10) have also been engineered for diagnostics, either in combination with components of the class 2 system or with the native type III complex22,23.
The first CRISPR-based diagnostic methods that were developed largely used Cas9 variants, which recognize double-stranded DNA (dsDNA). The main principles of the many different Cas9-based approaches for sensing DNA that have been reported include guide-directed reconstitution of split proteins by catalytically inactive Cas9 partners24, Cas9-based destruction of PAM-containing sites25,26 and Cas9-induced unwinding of the non-targeted DNA strand as a targeting site for isothermal amplification27. The Cas9-based method referred to as NASBACC (for nucleic acid sequence-based amplification (NASBA)-CRISPR cleavage)25 combines nucleic acid sequence-based amplification28 for the isothermal preamplification of targets, Cas9 cleavage for PAM-dependent target detection and a toehold sensor for the readout (Fig. 1, left). In brief, a toehold trigger is attached to a NASBA-amplified RNA fragment through reverse transcription. If a PAM sequence is present in the RNA fragment, Cas9-mediated cleavage leads to a truncated RNA without the trigger sequence. In the absence of the PAM sequence, the trigger containing full-length RNA activates the toehold switch, as indicated by a colour change. By sensing strain-specific PAM sites, the method allows for viral-lineage discrimination, as shown for the detection of the Zika virus (ZIKV) in infected monkey plasma at concentrations in the low femtomolar range25. Reported most recently, a Cas9-based method referred to as LEOPARD (for leveraging engineered tracrRNAs and on-target DNAs for parallel RNA detection) enables the multiplexed detection of different RNA sequences with single-nucleotide specificity29. The method is based on the observation that trans-activating crRNAs (tracrRNAs)—which in type II systems are necessary for the formation of Cas9–crRNA complexes—hybridize to cellular RNAs, making ‘non-canonical’ crRNAs. By reprogramming tracrRNAs to bind cellular transcripts of interest while allowing for the formation of Cas9–crRNA complexes, the resulting non-canonical crRNAs enable the guiding of Cas9 to a DNA target.
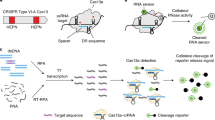
Left: RNA targets are amplified through NASBA, which starts with reverse transcription (RT) to complementary DNA using a sequence-specific primer that appends a trigger sequence (magenta) for the toehold sensor. The RNA from the RNA/DNA hybrid is destroyed by RNase H, enabling a primer that contains a T7 promoter to bind and create a complementary second DNA strand. T7 transcription of the dsDNA template creates the target RNA sequence, which can be used as starting material for a new NASBA cycle or detected by the toehold sensor. If a PAM sequence (blue) is present in the dsDNA amplicon, Cas9-mediated cleavage leads to a truncated template for T7 transcription, which generates a shorter target RNA that cannot activate the toehold sensor. In the absence of the PAM sequence, a full-length target RNA containing the trigger is transcribed, which activates the toehold sensor and produces a visible change in colour. Right: DNA or RNA are amplified by RPA or RT-RPA, respectively. For RNA-targeting CRISPR enzymes (including Cas13a), the amplified RPA product is T7-transcribed into RNA. Binding of the crRNA to the complementary target sequence activates the Cas enzyme and triggers collateral cleavage of quenched fluorescent reporters. Thereby, Cas13a (used in SHERLOCK) or Cas12a (used in DETECTR) indicate the presence of RNA or DNA target sequences, respectively.
The main difference between CRISPR type II (Cas9) systems and type V (Cas12) and type VI (Cas13) systems is the ability of the latter two systems to trigger non-specific collateral cleavage (trans cleavage) on target recognition. Collateral activity involves the cleavage of non-targeted single-stranded DNA (ssDNA; Cas12) or single-stranded RNA (ssRNA; Cas13) in solution, which enables the sensing of nucleic acids through signal amplification and allows for various readouts through the addition of functionalized reporter nucleic acids, which are cleaved by collateral activity. The CRISPR–Cas type VI (Cas13) family of enzymes has a size range of ~900–1,300 amino acids, detects ssRNA in cis conformation and shows collateral trans-cleavage activity against ssRNA in vitro30,31. In the Cas13-based assay SHERLOCK (for specific high-sensitivity enzymatic reporter unlocking)32, DNA or RNA is first isothermally amplified with recombinase polymerase amplification (RPA)33 or reverse transcription RPA (RT-RPA), respectively, using a forward primer that adds a T7 promoter to the amplicon (Fig. 1, right). This promoter allows for RNA transcription of the target, which is then recognized and bound by a complex of Cas13a from Leptotrichia wadeii (LwaCas13a) and a crRNA that includes a complementary sequence to the target. The activated Cas13 will then cleave both the on-target RNA by cis cleavage and, in a target-dependent manner, the ssRNA reporter molecules by collateral trans cleavage. The ssRNA reporter molecules consist of a fluorophore and a quencher joined together by a short RNA oligomer, which, once cleaved, allows for the separation of the fluorophore from the quencher, resulting in fluorescence. These processes enable the detection of viral RNA, bacterial DNA, human single-nucleotide polymorphisms (SNPs) and cancer-associated mutations with attomolar (10–18 M) sensitivity. Version two of the assay (SHERLOCKv2) enabled the quantitative multiplexed sensing of nucleic acids and the target detection at zeptomolar (10–21 M) concentrations, and introduced a lateral-flow readout based on an immunochromatographic assay, where cleaved reporter molecules are detected through antibody-conjugated gold nanoparticles on a paper strip22,34.
Cas12 enzymes that have been used in CRISPR-based diagnostics target dsDNA and ssDNA, require a PAM site in the target region for dsDNA cleavage and collaterally cleave ssDNA35. One of the first Cas12-based detection methods was reported in 2018 and referred to as DETECTR (for DNA endonuclease-targeted CRISPR trans reporter; Fig. 1, right)36. In this method, Cas12a from Lachnospiraceae bacterium (LbCas12a) or other organisms is guided to dsDNA targets by a complementary crRNA, triggering collateral cleavage of short ssDNA reporters carrying a fluorophore and a quencher. Similar to SHERLOCK, target recognition and reporter cleavage lead to the separation of the quencher from the fluorophore, which generates a fluorescence signal. DETECTR reached attomolar sensitivity when combined with RPA preamplification. Other Cas12-based techniques include HOLMES (for one-hour low-cost multipurpose highly efficient system)37,38, which employs PCR as preamplification together with LbCas12a, and HOLMESv239, which uses loop-mediated isothermal amplification (LAMP)40 combined with a thermostable Cas12b from Alicyclobacillus acidoterrestris (AacCas12b) in a one-pot reaction. Both HOLMES and HOLMESv2 showed a limit of detection (LOD) around 10 aM. Similarly, Cas12f targets dsDNA and ssDNA, but enables better discrimination of SNPs in ssDNA than Cas12a41.
Preamplification-free diagnostics
When using Cas enzymes without upstream preamplification of the target, most CRISPR-based diagnostics have a reported LOD in the picomolar range32,42. This LOD allows for target detection when there is a relatively high concentration of DNA or RNA in the sample. For example, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is apparently shed at high concentration in the early phase of infection (~6.76 × 105 copies per swab)43, allowing preamplification-free detection44. In addition to highly concentrated microbial samples, human genomic DNA (gDNA)45 and highly expressed human messenger RNAs30 and microRNAs (miRNAs)46 have been reported to be amenable to detection without preamplification.
To increase the sensitivity of preamplification-free CRISPR-based diagnostics, LwaCas13a has been combined with the CRISPR type III RNA nuclease Csm622. The nuclease activity of Csm6 is activated by cyclic oligoadenylates (2′,3′-cyclic phosphate groups)47,48. Because the collateral-cleavage activity of both LwaCas13a and PsmCas13b (from Prevotella sp. MA2016) generate products with hydroxylated 5′ ends and 2′,3′-cyclic phosphate ends, it was hypothesized that the collateral activity of Cas13 could generate Csm6 activators, thereby enabling amplified signal detection. In contrast to the cleavage preferences of LwaCas13a for the UU and AU two-base motifs, it was found that Csm6 from Enterococcus italicus (EiCsm6), Lactobacillus salivarius (LsCsm6) and Thermus thermophilus (TtCsm6) had strong cleavage preferences for A-rich and C-rich reporters, allowing for the cleavage activity of LwaCas13a and Csm6 to be independently measured in different channels and for the further design of ‘protected’ RNA activators. These activators were designed to contain a poly(A) Csm6 activator followed by a poly(U) stretch, on the basis of the rationale that the collateral cleavage of LwaCas13a would only degrade the uridines, thereby generating hexadenylates with 2′,3′-cyclic phosphate ends and activating Csm6. Activated Csm6 would in turn cleave additional reporter molecules, freeing up quenched fluorophores and thereby increasing the signal. This design led to a 3.5-fold increase in signal sensitivity when compared with Cas13a alone and when reading Csm6-mediated fluorescence and Cas13a-mediated fluorescence in the same channel. Similarly, the generation of cyclic oligoadenylates from the native Cas10 enzyme in type III systems has been used for detection of SARS-CoV-2 genomes23.
More strategies that increase the sensitivity of preamplification-free CRISPR-based diagnostics are needed. This could include the Cas-enzyme-mediated degradation of aptamers or of enzyme-inhibiting nucleic acid linkers (such as calf intestinal alkaline phosphatase or luciferase49) that can then be detected via colorimetric or luminescence methods.
Preamplification strategies
In contrast to applications that are amenable to preamplification-free CRISPR-based diagnostics, most clinical settings require the ability to sense nucleic acid concentrations below the picomolar range50. An example is the sensing of human immunodeficiency virus (HIV) or hepatitis C virus (HCV) load under antiviral therapy51,52. Therefore, CRISPR-based diagnostics reported so far largely rely on the preamplification of the target. Although some studies use PCR37,38, isothermal amplification techniques are more compatible with POC use, owing to simpler instrumentation requirements (Table 3). Some of these methods function at near-ambient temperature (in particular, RPA, 37–42 °C), but others require heating devices (in particular, NASBA, 40–55 °C; LAMP, 60–65 °C; strand-displacement amplification53 (SDA), 60 °C; helicase-dependent amplification54 (HDA), 65 °C; and exponential amplification reaction55 (EXPAR), 55 °C). In CRISPR-based diagnostics, the most commonly used isothermal amplification methods are RPA and LAMP. Initially, SHERLOCK32 and DETECTR36 used RPA for preamplification, which exhibited greater sensitivity than NASBA32. Although non-specific amplification represents a challenge in conventional RPA, the downstream crRNA-based target detection improves specificity. The design of RPA primers is less complex than the design of LAMP primers, but reliable procurement of the proprietary enzyme mix of a single supplier has been challenging. Furthermore, the addition of macromolecular crowding agents (such as poly(ethylene glycol)) together with low reaction temperatures reduces the mixing of the reagents and requires a mixing step for the amplification of lowly abundant targets56.
One-pot detection by using HOLMESv2 is possible by combining AacCas12b with LAMP39. During the COVID-19 pandemic, reverse transcriptase LAMP (RT-LAMP) has been widely used as an isothermal preamplification technique. With RT-LAMP, a higher operating temperature than used in RPA enables increased specificity of amplification, and the commercial availability of the necessary enzymes and reagents facilitates the planning and performing of experiments. The design of LAMP primers can be complex, but online tools (https://primerexplorer.jp and https://lamp.neb.com) can help implement this amplification technique.
Quantification strategies
In many clinical settings, an accurate quantification of the biomarker concentration is desirable, especially if changes indicate treatment efficacy such as during the monitoring of viral load under antiviral therapy57. For PCR, quantification can be absolute or relative. In absolute quantification, serial dilutions of an external standard with known concentrations are performed to obtain a standard curve, which is used to determine the target concentration in the unknown sample58. In relative quantification, which is often used to determine differentially expressed genes, the target concentration is expressed in relation to a reference59.
In CRISPR-based diagnostics, absolute quantification through comparison with a standard curve can be achieved within the picomolar-to-micromolar range (10–12 M–10–6 M), where CRISPR-based collateral-cleavage activity correlates with target concentration32. However, clinically useful assays generally require a lower LOD and thus preamplification. In a two-pot reaction, saturation during preamplification can prevent a quantitative readout32. For one-pot reactions, where preamplification and CRISPR-based detection are combined, collateral cleavage happens simultaneously with target amplification22,34,39,60,61,62,63. This enables a real-time readout that is amenable for quantification when a continuous measurement of the signal is incorporated. However, it requires more sophisticated laboratory equipment (similar to qPCR), which is usually not available for POC tests. To facilitate quantification in two-pot reactions, lower RPA-primer concentrations can be used to prevent saturation of the preamplification reaction22. In the SHERLOCKv2 protocol, a 240 nM primer concentration allows for the correlation of RNA input concentration and output signal within the attomolar-to-picomolar range (10–18 M–10–12 M). Additional options for quantification include the possibility of serial dilutions of the unknown sample compared with a known standard. The implementation of these different quantification solutions with POC-amenable devices will probably be a major focus of research in CRISPR-based diagnostic technologies. Also, controlling for the quality of nucleic acid preparations and for any inhibitory components introduced during sample collection or processing (such as heparin, excess salts, urea, haem, ionic detergents, alcohols and phenols) remains important for CRISPR-based diagnostics to reach the clinical utility of current diagnostic PCR protocols64.
Strategies for the detection of multiple targets
Comprehensive and simultaneous testing of multiple targets (multiplexing) is desirable in many diagnostic settings, as it can provide more information at higher throughput and at a lower cost per target5. Although sequencing technologies enable multiplexing through the addition of unique identifiers that are deconvoluted during data analysis65, PCR-based nucleic acid detection can be multiplexed through the simultaneous amplification of different targets in one reaction combined with the specific detection of each amplicon66, or through the parallel readout of multiple separate reactions.
Initial efforts in the design of multiplexed CRISPR-based diagnostics took advantage of the specificity of different Cas enzymes with regards to their collateral-cleavage activity towards different nucleic acid reporter sequences. This allowed multiple nucleic acid targets to be detected by Cas enzymes within one assay22. In particular, SHERLOCKv2 enabled the qualitative detection of up to four different targets in one multiplexed reaction by including four reporter-molecule designs that were specific to the orthogonal collateral-cleavage base preferences of PsmCas13b, LwaCas13a, CcaCas13b (Capnocytophaga canimorsus Cc5) and AsCas12a (from Acidaminococcus sp.). Multiplexed Cas13 detection could be combined with multiplexed preamplification (using RPA), allowing for the simultaneous detection of two different targets (Fig. 2, left). Additional multiplexing beyond these levels in a single reaction was limited by the preamplification chemistries22.
Left: pooled multiplexing of up to four targets, as implemented in SHERLOCKv2. Orthogonal CRISPR enzymes (PsmCas13b, LwaCas13a, CcaCas13b, AsCas12a), each cleaving preferentially different reporter molecules bearing different fluorophores (FAM, TEX615 (TEX), Cy5, hexachlorofluorescein (HEX)) with distinct absorbance and emission wavelengths. Right: in CARMEN, a distinct colour code is added to each PCR-amplified sample or Cas13-based detection mix. The Cas13-based detection mix contains Cas13a, a sequence-specific crRNA and a cleavage reporter. The colour-coded solutions are emulsified in fluorous oil, which creates nanolitre droplets. Droplets from all samples and detection mixes are pooled together and loaded into a microwell-array chip in one step to create all possible pairwise combinations. The droplet pair in each well is identified using fluorescence microscopy before they are merged by exposure to an electric field. Fluorescence microscopy is then used to monitor each Cas13-based detection reaction.
The CARMEN (for combinatorial arrayed reactions for multiplexed evaluation of nucleic acids) CRISPR-based multiplexed assay enables the scalable and parallel detection of more than 4,500 targets67. The method builds on the reaction chemistry of SHERLOCK, yet uses a miniaturized reaction volume (Fig. 2, right). In CARMEN, amplified nucleic acid target samples and LwaCas13 detection mixes are each combined with a distinct solution-based colour code. The colour-coded solutions are emulsified in fluorous oil, creating nanolitre droplet inputs. The droplets from all of the samples and detection mixes are pooled and subsequently loaded onto a microfabricated chip to create all possible pairwise combinations of sample and detection droplets (one pair per well). Droplet pairs are then merged by applying an external electric field. An initial recording of the dyes of two droplets in a particular microwell indicates which sample and detection droplet are present, and fluorescence generated through Cas13a-triggered reporter cleavage on target detection indicates a positive reaction. This miniaturization coupled with the high sensitivity and specificity of Cas13a enabled the detection of 169 human-associated viruses, the subtyping of coronaviruses and influenza A strains, and the identification of HIV mutations that are drug resistant. Compared with traditional DNA microarrays, CARMEN does not require the time-consuming spotting of probes and offers higher scalability than PCR panels. Although it requires a preamplification step, microscopy analysis and skilled lab personnel, CARMEN allows for fast, high-throughput and low-cost pathogen detection, which is critically needed in clinical routines and for the surveillance of emerging public-health threats. Owing to the miniaturized reactions for detection, CARMEN considerably reduces reagent costs and sample consumption. However, its current dependence on advanced microscopy limits the use of the technology to resource-rich laboratory settings.
Combination of CRISPR–Cas systems with sequencing
Because sequencing-based diagnostic technologies use the alignment of reads to bioinformatic databases for pathogen identification, they tolerate variability in the genome of the target organism and do not necessarily require prior knowledge of the target sequence. These features are particularly useful when the pathogen is unknown, or when its genome evolves quickly. However, to detect low-abundance pathogens, the enrichment of regions of interest before sequencing becomes necessary. PCR-based amplification of sequencing regions has several limitations, including the removal of epigenetic marks, amplification bias and challenges associated with the amplification of GC-rich regions or of very large regions. Larger amplicons are of particular interest for long-read sequencing techniques, such as nanopore sequencing, because they allow for the characterization of long-range structural variations.
To overcome these limitations, several protocols use Cas nucleases to enrich regions of interest before sequencing. For example, nanopore Cas9-targeted sequencing (nCATS) uses Cas9 to cleave chromosomal DNA for the ligation of adapters for nanopore sequencing (Fig. 3, left), enabling the targeted sequencing of long fragments at high depth while maintaining epigenetic marks68. This technology allowed the simultaneous detection of CpG methylation, structural variations and haplotype-resolved single-nucleotide variants using 3 μg of genomic DNA, and it achieved 675× coverage with a MinION flow cell and 34× coverage with the Flongle flow cell (both cells are commercialized by Oxford Nanopore Technologies). In a similar effort to circumvent the need for PCR-based enrichment, STRique (for short tandem repeat identification, quantification and evaluation) was developed69 for the identification of short tandem repeats (STRs) with nanopore sequencing (Fig. 3, left). In addition to Cas9, Cas12a was used to induce DNA cuts next to a repeat-containing region, followed by the ligation of adapters for nanopore sequencing. Using an algorithm that identified the positions of the STR-flanking regions and the number of STRs between them, the method enabled the detection of STR expansion alongside CpG methylation status in cell lines derived from patients with fragile X syndrome and with amyotrophic lateral sclerosis.
Left: nCATS is a Cas9-based enrichment technology, which first dephosphorylates DNA ends and then uses Cas9 to induce cuts near a region of interest (ROI) that contains a PAM sequence. A DNA polymerase I with an N-terminal truncation that lacks exonuclease activity (Klenow fragment) enables the addition of dAMP (deoxyadenosine monophosphate (dA)) to the 3′ end the DNA fragment (d(A)-tailing)). Next, adapters containing a motor protein are ligated to the DNA at Cas9 cleavage sites, enabling the DNA to pass through the nanopore. STRique uses Cas12a for the enrichment of repeat-containing regions analysed by nanopore sequencing, and employs an algorithm for the identification of the position of the repeat flanking regions and the number of repeats. Right: DASH uses Cas9 to deplete unwanted sequences (black) before next-generation sequencing. Non-targeted sequences (magenta) retain intact adapters for amplification and sequencing.
CRISPR–Cas systems have also been combined with targeted next-generation sequencing to increase the sequencing yield. For example, DASH (for depletion of abundant sequences by hybridization) was developed70 to enrich pathogen sequences and to remove unwanted species. In this method, a library of crRNAs directs Cas9 to unwanted target sequences for cleavage, and non-targeted regions retain the intact adapters required for amplification and sequencing (Fig. 3, right). Another method, FLASH (for finding low-abundance sequences by hybridization), includes the blocking of gDNA by phosphatase and digestion through Cas9 complexed to a set of crRNAs targeting genes of interest, followed by the ligation of adapters, and by amplification and sequencing71. This technique detected, in respiratory fluids and dried blood spots, subattomolar levels of drug-resistant Gram-positive bacteria and the malaria parasite Plasmodium falciparum.
Assay optimization for applications at the POC and in the field
Readouts
Some strategies for measuring and visualizing the activation of Cas enzymes on target recognition have employed fast and low-cost readouts amenable for POC or field applications. The usage of reporter molecules with different properties enables a wide variety of readouts, including fluorescence, lateral-flow immunochromatography and electrochemistry. For fluorescence-based assays, most CRISPR-based diagnostic methods take advantage of RNA or DNA reporter molecules carrying a fluorophore and a quencher. Collateral-cleavage activity on target detection causes the spatial separation of the fluorophore and the quencher, and the resulting fluorescence can be read on conventional plate readers (Fig. 4, top left), on low-cost and portable devices72 and even with the naked eye under blue light60 (Fig. 4, top left). Additional strategies enable colorimetric visualization by using gold nanoparticles, which are linked through ssRNA or ssDNA oligonucleotides. Cas12-mediated or Cas13-mediated trans cleavage of the linker resulted in the dispersal of the gold nanoparticles on target recognition, and produced a vibrant red colour in solution73. Alternatively, Cas-mediated cleavage of biotinylated ssDNA substrates resulted in a decrease in the magnetic pull-down of DNA gold nanoparticles74. A turbidity-based assay based on the liquid–liquid phase separation of nucleic acids and positively charged polyelectrolytes can also provide visual readouts75. In this case, collateral cleavage leads to the degradation of nucleic acid polymers on target recognition. This can be visualized after complementation with polycations, where the solution either stays clear when cleavage has occurred or becomes turbid in the absence of target-triggered cleavage.
Top left: fluorescence readouts rely on quenched fluorophores that, on target recognition, are released as a result of the collateral cleavage of the DNA or RNA oligomers joining the fluorophore and quencher. The signal can be measured by a fluorescence reader or read by eye under blue light. LED, light-emitting diode. Top right: lateral-flow-based immunochromatographic readouts rely on the high affinity of streptavidin and biotin, and use a reporter molecule consisting of a DNA or RNA oligomer conjugated to FAM and biotin. The product of the CRISPR reaction is deposited on the sample-application area of a test strip, at which point FAM binds to gold-nanoparticle-labelled FITC-specific antibodies. When intact, the reporter oligomer remains bound to the streptavidin line through biotin, creating one colour band on the test strip. When the reporter oligomer is cleaved, FAM and the bound gold-labelled FITC-specific antibodies flow farther on the strip and bind to secondary anti-species antibodies, which leads to the formation of a second colour band. Bottom left: electrochemical readout relying on the amperometric detection of H2O2 in an electrochemical cell46. Anti-fluorescein antibodies (green) coupled with glucose oxidase (GOx) bind to FAM-carrying RNA reporter molecules, which are immobilized through biotin on a surface. The concentration of GOx is reduced in the presence of an RNA trigger, owing to the activated crRNA/Cas13a complex cleaving the RNA reporter molecule on target recognition, which frees the GOx-carrying anti-fluorescein antibodies from the surface. Bottom right: in the presence of a dsDNA trigger, Cas12a collaterally cleaves ssDNA linkers within a hydrogel. This prevents crosslinking within the hydrogel, thereby modulating the permeability of a paper-based microfluidic device and allowing for buffer flow, which can be measured electrically79,80.
Lateral-flow assays (Fig. 4, top right) offer a simple visualization of the collateral cleavage activity inherent to many CRISPR-based diagnostics. A commercially available system (Millenia 1T) first reported in ref. 22 has been widely adopted. In this technique, reporter molecules carrying biotin and fluorescein (fluorescein amidite (FAM)) bind to anti-fluorescein isothiocyanate (FITC) antibodies coupled to gold nanoparticles in the sample pad area of the strip. These complexes subsequently travel to a streptavidin-capture line, where they bind through the biotin of the reporter molecule. In the absence of the target, uncut reporter molecules are retained there, and this is visualized as one band on the strip. In the presence of the target, Cas-enzyme-mediated collateral cleavage of the reporter separates the biotin from the FAM molecules bound to the anti-FITC gold nanoparticles, allowing them to travel farther on the strip and generating a second visible band at the antibody-capture line, which bears species-specific secondary antibodies. An alternative to collateral cleavage-based target detection combines Cas9-based amplicon detection with a lateral-flow readout using a crRNA-anchoring-based hybridization assay76.
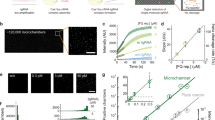
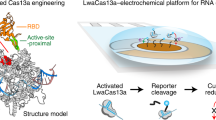
CRISPR-based diagnostics with electronic readouts are also possible. One method, named CRISPR–Chip, involved deactivated Cas9 complexed with a target-specific crRNA and immobilized on a graphene-based field-effect transistor45. On target binding, alterations to graphene’s conductivity caused the electrical properties of the transistor to change, which can be measured as a change in current. Notably, this technology did not require labelled reporter molecules and could sense deletions in the dystrophin gene in unamplified gDNA samples from patients with Duchenne muscular dystrophy. The sensitivity of the system reached the femtomolar range (~1.7 fM). Another strategy for obtaining electronic readouts, named E-CRISPR, involved an ssDNA-cleavage reporter with a methylene blue electrochemical tag and a thiol moiety for immobilization on a gold electrode77. In this method, Cas12a-mediated collateral cleavage upon target detection led to a decrease in the electrochemical current through the release of methylene blue, whereas the absence of target resulted in a high electrochemical current, owing to an intact methylene blue-carrying reporter. This assay detected human papillomavirus and parvovirus B19 DNA with a LOD of ~50 pM, and can be adapted for protein sensing. Another method used an electrochemical biosensor to detect Cas13a-mediated collateral cleavage of RNA reporters carrying FAM and biotin46 (Fig. 4, bottom left). In this approach, the binding of glucose-oxidase-coupled anti-FAM antibodies to reporter molecules immobilized on the sensor catalysed the oxidation of glucose. The produced H2O2 was then amperometrically detected by using an electrochemical cell with a platinum working electrode, a platinum counter electrode and a silver/silver chloride reference electrode. This technique allowed the detection of miRNAs at a LOD of 10 pM within 4 hours in volumes smaller than 0.6 μl (this LOD was achieved using a separate ‘off-chip’ cleavage; the all-in-one sensor had a LOD of 2.2 nM). Cas13a-based electrochemical detection of miRNAs was also shown with a portable electrochemiluminescence chip78 (with a reported limit of detection of 1 fM). In this technique, the miRNA-mediated activation of Cas13a led to the collateral cleavage of a trigger for the initiation of exponential isothermal amplification. The amplified products formed a complex with [Ru(phenanthroline)2dipyridophenazine]2+, and the oxidation of the complex at the anode of a bipolar electrode generated an electrochemiluminescent signal proportional to the concentration of the target miRNA.
DNA hydrogels have been designed to enable CRISPR-based diagnostic readouts through changes in their material property79,80. The hydrogels consist of water-filled polymers containing DNA molecules serving as an anchor or structural element within the gel (Fig. 4, bottom right). On target recognition, Cas12a triggers collateral cleavage of the DNA molecules, which changes the properties of the gel. This technology was used for sensing pathogen-derived nucleic acids in a multilayered paper-based microfluidic device (μPAD) with dual visual and electronic readouts. In the absence of the target, the DNA-linker-containing hydrogel obstructed the flow of a buffer through the porous channels of the paper-based device. Target recognition led to Cas12a-mediated linker destruction, which prevented hydrogel crosslinking and caused visually observable flow through the porous μPAD channels. This technique allowed for the detection of dsDNA down to 400 pM without preamplification. To create an electronic readout, two electrodes were placed in the bottom layer of the μPAD. Because the electrical conductivity of the gel depended on its degree of crosslinking, low levels of crosslinking due to Cas12a-mediated cleavage caused an electrolyte-containing buffer to flow, whereas high levels of crosslinking in the absence of any target blocked the buffer from flowing through, thereby preventing the recording of an electric signal. Moreover, the incorporation of a wireless radiofrequency identification (RFID) module into the μPAD enabled the monitoring of target detection at different geographic locations in real time. To this end, Cas12a-mediated collateral cleavage on target detection resulted in the short-circuiting of an interdigitated electrode arrangement in the loop RFID tag, which increased the strength of the received signal with respect to that of a reference RFID tag.
Sample preparation
Sample processing is a critically important consideration for improving the usability of CRISPR-based diagnostics for field applications and POC settings. CRISPR-based diagnostics initially used synthetic targets or time-consuming or equipment-intensive nucleic acid isolation protocols. More recent efforts aim for the rapid and equipment-free isolation of DNA or RNA. Importantly, the reagents used in the sample-extraction steps must not interfere with any downstream preamplification steps or with CRISPR-based target detection. As in many cases, the final readout is based on collateral-cleavage activity, and special precautions must be taken to ensure the absence of RNases and DNases, which can cleave ssRNA or ssDNA reporters.
The original SHERLOCK protocol showed that saliva could be processed directly by using 0.2% Triton X-100 in a lysis mixture with phosphate buffered saline, and by heating at 95 °C for 5 min. This crude saliva mixture was used in a SHERLOCK reaction for genotyping. To expand to additional sample types, the protocol HUDSON (for heating unextracted diagnostic samples to obliterate nucleases) enabled the inactivation of nucleases and viruses through heat and chemical reduction through the addition of tris(2-carboxyethyl)phosphine hydrochloride at a 100 mM final concentration and ethylenediaminetetraacetic acid (EDTA) at a 1 mM final concentration81. Different heating steps (ranging from 5 min to 20 min and from 37 °C to 95 °C) were optimized for different targets and sample types. HUDSON enabled the detection of RPA-preamplified targets through CRISPR–Cas13a, including the sensing of ZIKV and dengue virus in blood, plasma, serum, saliva and urine. Since the HUDSON protocol was not sufficient to inactivate nucleases for the detection of Plasmodium species (as observed by the fraction of false-positive samples), a preparation buffer containing stronger chelating agents has been developed (20% w/v Chelex100 TE-buffer containing dithiothreitol (DTT) at 50 mM)61.
A commercially available fast DNA-extraction solution (QuickExtract, Lucigen) enables fast RNA preparation for the detection of SARS-CoV-2 RNA through RT–qPCR82. Here nasopharyngeal swabs are mixed with the extraction solution, incubated at 95 °C for 5 min and directly used in the RT–qPCR reaction after cool down. This isolation protocol is compatible with downstream RT-LAMP preamplification and detection through Cas12b from Alicyclobacillus acidiphilus (AapCas12b)63. QuickExtract also enabled the lysis of samples at 22 °C or 60 °C for 10 min, and when proteinase K was blocked in the downstream reactions by an inhibitor.
Large-scale testing and costs
One advantage of CRISPR-based diagnostic assays is their independence from complex and costly laboratory equipment and from common reagents such as PCR master mixes. Because Cas enzymes and crRNAs can be produced fast and at scale, CRISPR-based assays are less dependent on supply-chain issues. However, although the costs of buffers, primers and the oligos used as templates for crRNA synthesis are negligible, CRISPR-based assays include many more enzymes than PCR, especially when an isothermal preamplification step is included. The cost for an individual CRISPR-based diagnostics reaction with preamplification is approximately US$0.61 per reaction in an experimental setting32. Of these costs, RPA reagents made up the largest fraction. However, the first commercially available CRISPR-based diagnostic assay for SARS-CoV-2 including RT-LAMP as preamplification is currently available at US$30.15 per reaction (https://www.idtdna.com/pages/landing/coronavirus-research-reagents/sherlock-kits). It is unclear how prices will develop after the approval of more tests, and how patents that cover methods for the sensing of nucleic acids through CRISPR–Cas proteins will affect the dissemination of the technology.
Most CRISPR-based diagnostic assays use liquid reagents, which afford experimental flexibility. However, the storage of solutions of the crRNA, cleavage reporter and Cas protein typically requires freezers that maintain ultralow temperatures. Lyophilization of these components and of the preamplification reagents eliminates the dependence on cold chains, and by using higher input volumes of sample the assay sensitivities can be similar31 or even higher61 than with the use of non-lyophilized reactions. Systematic stability testing of the components for CRISPR-based diagnostics is needed, yet studies in clinical cohorts83 and the assessment of daily experimental variation84 suggest that the results of the assays are highly reproducible.
Biomedical applications
CRISPR-based diagnostics have been used for a wide range of biomedical applications, and in particular for the sensing of nucleic-acid-based biomarkers of infectious and non-infectious diseases and for the detection of mutations and deletions indicative of genetic diseases. Moreover, the technology has been adapted for the sensing of proteins and small molecules.
Infectious diseases
The main focus of CRISPR-based diagnostics has been the detection of pathogens; in particular, the DNA or RNA of viruses, bacteria and parasites. RNA viruses sensed with CRISPR-based methods include parvovirus B19 (ref. 77), members of the family Flaviviridae (dengue22,25,81, Zika25,32 and Japanese encephalitis virus37), Ebola85 and Coronaviridae (which have naturally become a major focus of interest during the COVID-19 pandemic).
The SHERLOCK-based detection of SARS-CoV-2 initially consisted of a two-step assay comprising RPA-based preamplification, followed by T7 transcription and Cas13a-mediated target recognition86. This assay has been tested in a qPCR-validated clinical cohort containing 154 samples analysed by lateral-flow and fluorescence readouts, and with 380 PCR-negative samples analysed by fluorescence only83. The assay reached a LOD of 42 RNA copies per reaction, and sensitivities of 96% and 88% when using fluorescence and lateral-flow readouts, respectively. Both readouts were 100% specific. In May 2020, a modified SHERLOCK-based two-step assay using RT-LAMP instead of RT-RPA received emergency use authorization from the United States Food and Drug Administration for the CRISPR-based detection of SARS-CoV-2. Testing with this assay is intended for the qualitative detection of SARS-CoV-2 in specimens of the upper respiratory track, and is limited to laboratories authorized by CLIA (for clinical laboratory improvement amendments) regulations. A different Cas13a-based assay, SHINE (for SHERLOCK and HUDSON integration to navigate epidemics)62, combined the preamplification reaction and CRISPR-based detection into a single-step reaction that uses HUDSON to accelerate SARS-CoV-2 viral extraction in nasal swabs and saliva samples. The addition of RNase H, the optimization of buffers, concentrations of magnesium and of primers, and the usage of a polyU reporter together with a SuperScript-IV reverse transcriptase, reduced the LOD to a range similar to that of the two-step assay (10 copies per μl with the fluorescent readout, and 100 copies per μl with the lateral-flow readout). The test was validated on 50 nasopharyngeal patient samples, and achieved 90% sensitivity and 100% specificity against RT–PCR. In a different Cas13-based approach, the integration of multiple crRNAs, the usage of a Cas13a homologue from Leptotrichia buccalis (Lbu) and the measurement of fluorescence over time enabled the detection of SARS-Cov-2 RNA extracted from nasal swabs down to 1.27 × 108 copies per ml (1.65 × 103 copies per μl in the Cas13 reaction). Importantly, this was achieved without the need for preamplification, and the assay was compatible with readout using a portable fluorescence-detection device44.
By using Cas12a, the two-step assay SARS-CoV-2 DETECTR was optimized to detect the N (nucleoprotein) and E (envelope small membrane protein) genes of the SARS-CoV-2 genome, as well as a human control gene (for the ribonuclease P protein), in less than 40 min from respiratory swab RNA extracts preamplified by RT-LAMP87. SARS-CoV-2 DETECTR returned results with a 95% positive predictive agreement and a 100% negative predictive agreement relative to the RT–qPCR assay from the United States Centers for Disease Control and Prevention. Cas12a was also used in the SARS-CoV-2 AIOD-CRISPR (for all-in-one dual CRISPR–Cas12a) assay60, which uses Cas12a together with a pair of non-PAM-dependent crRNAs, each sensing separate regions of the target nucleic acid amplicon. These Cas12a-crRNA complexes were introduced into a one-pot assay containing the reagents for RPA-based preamplification and the cleavage reporter. This approach enabled the detection of 5 copies of synthetic SARS-CoV-2 RNA per reaction, and was validated in 28 clinical swab samples. STOPCovid (for SHERLOCK testing in one pot)63, another Cas12-based one-pot assay, integrated RT-LAMP-based preamplification with CRISPR-mediated detection in a one-step workflow. Of the various Cas proteins explored, Cas12b from Alicyclobacillus acidiphilus (AapCas12b) together with the scaffold single guide RNA from AacCas12b, allowed the test to run at the temperature range required for LAMP. STOPCovid has a sensitivity comparable to that of RT–qPCR, with a LOD of 100 copies of viral genome per reaction.
Besides RNA viruses, DNA viruses such as Herpesviridae (including cytomegalovirus (CMV)84 and Epstein–Barr virus88), Polyomaviridae (BK virus (BKV))84 and Papillomaviridae (human papillomavirus)36 have also been detected with CRISPR-based diagnostics. In particular, a SHERLOCK-based assay was used to detect CMV and BKV in a clinical setting84.The assay was validated with qPCR, and detected BKV from 67 HUDSON-isolated plasma and urine samples with 100% sensitivity and 100% specificity. HUDSON-based processing of CMV-containing plasma samples resulted in 80% sensitivity and 100% specificity, whereas DNA purification through silica-based membranes was necessary to reach 100% sensitivity and 100% specificity. To objectively assess the results of the lateral-flow assay, a smartphone-based software application was developed to quantify band intensities. The background signal of the lateral-flow assay was independent of daily experimental variation, incubation time and temperature, which facilitated the reproducibility of the qualitative readout.
Bacteria detected with CRISPR include Mycobacterium tuberculosis32,89, Staphylococcus aureus22,32,90, Listeria monocytogenes91, Pseudomonas aeruginosa32 and Salmonella enteritidis92. CRISPR has also been adapted to sense the parasites of the Plasmodium group responsible for malaria. This includes the sensing of different P. falciparum strains71 and the development of a pan-plasmodium assay93 that detected all Plasmodium species known to cause malaria in humans as well as the species-specific detection of P. vivax and P. falciparum. A different CRISPR-based assay based on an RPA–Cas12a one-pot reaction using lyophilized reagents downstream of a simplified sample preparation was developed for the detection of P. falciparum, P. vivax, P. ovale and P. malariae61. The addition of a reverse transcriptase to transcribe target RNA into DNA increased the assay’s sensitivity. The protocol achieved a LOD between 0.36 and 2.4 parasites per μl, depending on the Plasmodium species.
Non-infectious diseases
CRISPR-based diagnostics have also been developed for the detection of RNA species relevant to non-infectious diseases. For example, CRISPR-based sensing of human CXCL9 mRNA was used to detect acute cellular kidney-transplant rejection, as defined by renal biopsy, with a sensitivity of 93% and a specificity of 76%, in total RNA isolated from 31 urinary cell pellets84. CRISPR-based diagnostics have also been used to sense miRNAs, as exemplified by the CRISPR/LwaCas13a-based electrochemical detection of miR-19b in serum samples of patients with medulloblastoma46 without preamplification, and the testing of RNA isolated from breast cancer cell lines for miR-17 with CRISPR/LbuCas13a94 and for miR-21 with LbCas12a, after rolling-circle transcription95.
SNPs and deletions
A major strength of CRISPR-based diagnostics is the single-nucleotide specificity of Cas enzymes, which enables the detection of point mutations and small deletions. Single-nucleotide specificity has made possible the CRISPR-based detection of markers of antimicrobial resistance, of deletions and mutations in the epidermal-growth-factor-receptor gene22, of mutations conferring Duchenne muscular dystrophy45 and of SNPs in the E3 ubiquitin ligase gene conferring eye colour41. The single-nucleotide specificity of the CRISPR–Cas system has also been leveraged for the sensing of miRNAs, which are challenging to detect because of their short size and because they can differ by only a single base. CRISPR-based approaches may also be suitable for the detection of circulating SARS-CoV-2 variants that might differ in pathogenicity and transmissibility96.
Targets other than nucleic acids
CRISPR-based diagnostic methods have been adapted for the detection of proteins and small molecules. In these cases, the CRISPR system is primarily used as a reporter or amplifier, and the actual sensing of the target molecule is mediated through proteins or aptamers that undergo a conformational change on target recognition. One strategy for sensing small molecules combined CRISPR–Cas12a with bacterial allosteric transcription factors97 (aTFs). Referred to as CaT-SMelor (for CRISPR–Cas12a-mediated and aTF-mediated small molecule detector), the method was applied to sense uric acid and p-hydroxybenzoic acid at nanomolar concentrations, and thereby enabled the detection of uric acid in human blood samples. In this method, the incubation of immobilized aTF–dsDNA complexes containing the Cas12a target sequence with the aTF target small molecule led to a conformational change in the aTF that freed the dsDNA from the complex. After removal of the immobilized aTF by centrifugation, free dsDNA was detected using CRISPR–Cas12a and measured through the collateral cleavage of a quenched fluorescent reporter. SPRINT (for SHERLOCK-based profiling of in vitro transcription)98 used aTFs and riboswitches that are triggered by specific effector molecules to regulate transcription of RNA targets. Transcribed RNA is then detected by a crRNA and Cas13a. This enabled the sensing of a wide range of effector molecules (including fluoride, adenine, guanine, S-adenosylmethionine, flavin mononucleotide, serotonin, zinc and anhydrotetracycline).
A different approach to sensing small molecules involved an aptamer that contained an ssDNA target of Cas12a, which served as a sensor for adenosine triphosphate99 (ATP; reported LOD of the assay, 400 nM of ATP). In this approach, the binding of ATP to the aptamer hindered target recognition and thereby reduced the Cas12a-triggered collateral cleavage of an ssDNA reporter. Alternatively, ATP or Na+ ions have been sensed through aptamers or DNAzymes (also referred to as ‘functional DNA’) that release Cas12a targets on binding to their non-nucleic acid targets, resulting in the activation of Cas12a as detected by the trans cleavage of fluorescent reporter molecules100.
Similarly, an aptamer-based detection method has been developed for the CRISPR–Cas12a-based sensing of the transforming growth factor β1 (TGF‐β1) protein77. In this method, TGF-β1 binds to a specific ssDNA aptamer that contains a Cas12a target site. In the absence of TGF-β1, all free and unbound aptamer molecules are recognized by Cas12a, whose collateral-cleavage activity of ssDNA reporters that carry methylene blue results in a decrease of the electrochemical signal, which is sensed by E-CRISPR. The aptamer-based E-CRISPR assay allowed for the detection of TGF-β1 in clinical samples with a LOD of 0.2 nM. And CLISA (for CRISPR–Cas13a-based signal amplification linked immunosorbent assay) was developed to sense human interleukin-6 at 2.29 fM and human vascular endothelial growth factor at 0.81 fM (ref. 101). This was achieved by replacing the signal-amplification enzyme of a sandwich-type enzyme-linked immunosorbent assay (using horseradish peroxidase) with biotinylated dsDNA containing a T7 promoter, which allowed T7-mediated transcription of a Cas13a target, leading to its activation and to the collateral cleavage of a reporter.
Outlook
In less than five years, CRISPR-based diagnostics have evolved from an experimental nucleic acid sensing tool to a clinically relevant diagnostic technology for the fast, affordable and ultrasensitive sensing of biomarkers at the POC. However, there are several challenges that the technology needs to overcome to be able to deliver on the promise of transforming the way diseases are diagnosed and emerging pathogens monitored. A major shortcoming of most current CRISPR-based diagnostics is their dependence on preamplification for the detection of targets below the femtomolar range. Although the primers used for preamplification add another layer of specificity, this process adds complexity to the assay, increases its costs and prolongs the reaction time. The incorporation of non-primer-based signal-amplification strategies, or modifications of the Cas enzyme, crRNA or reporter molecule, are potential paths for improvement. Currently, most crRNAs and primers for preamplification have to be experimentally tested, and the design rules to narrow down potential candidates are coarse. Machine learning and bioinformatics algorithms based on large-scale experimental datasets could allow for the development of more effective design tools for the better prediction of crRNA performance and for improving crRNA specificity and activity. In fact, the feasibility of such approaches for crRNA design has been shown in the context of genome engineering102,103 and in the characterization and optimization of toehold switches104,105.
The ease of use of CRISPR-based diagnostics has been improved through optimized one-pot reactions and the simple visualization of test results. However, sample preparation still requires a separate step, and incubation temperatures higher than room temperature necessitate heating devices. In addition, target concentrations close to the LOD of the assay make it challenging to quantify the readout with certainty, especially when using the lateral-flow format. For usage at home or in the field, the assay design should combine simple sample-preparation protocols with robust detection methods to deliver robust results in variable or challenging contexts, such as prolonged storage, limited user training and harsh environmental conditions. To this end, devices that integrate sample preparation, sensing and reporting in single and easy-to-operate units are increasingly possible by using microfluidic techniques106. And the combination of such devices with digital analysis and data-sharing systems holds promise for patient-centric testing and remote data access. This is particularly important for mitigating the spread of infectious diseases by limiting unnecessary in-person contact in hospitals. Also, CRISPR-based diagnostics may enable patient-centric companion nucleic-acid-based diagnostics for monitoring treatment responses to a drug and to differentiate patients that are at risk for serious adverse effects from those that will probably benefit from the drug107. CRISPR-based diagnostics could facilitate the monitoring of genetic markers indicative of treatment response, such as mutations in the BRAF gene, which are commonly used to inform the treatment of melanoma skin cancer108. Furthermore, CRISPR-based diagnostics may be used for the real-time monitoring of gene expression across different tissues through the sensing of cell-free mRNA109.
Future applications of CRISPR-based diagnostics will probably involve technologies at the intersection with materials science79,80, for the sensing of nucleic acids on solid surfaces, on wearables and on medical devices. For example, ongoing work aims to incorporate SARS-CoV-2 sensors into face masks for the real-time detection of the virus. Direct sensing on surfaces can take advantage of high local concentrations of infectious agents and could allow for continuous tracking and for early diagnosis. This could extend the capabilities of monitoring strategies, such as the use of toilet systems for the analysis of excreta110.
Newly developed CRISPR assays must, of course, be validated83 by clinical trials, and assay validity monitored and maintained after clinical implementation. Yet, we believe that rapid innovation in CRISPR-based diagnostics will end up reshaping the technological landscape of nucleic-acid-based detection.
References
Neal, R. D. et al. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer https://doi.org/10.1038/bjc.2015.48 (2015).
Yang, S. & Rothman, R. E. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(04)01044-8 (2004).
Weissleder, R., Lee, H., Ko, J. & Pittet, M. J. COVID-19 diagnostics in context. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abc1931 (2020).
Burki, T. K. Testing for COVID-19. Lancet Respir. Med. https://doi.org/10.1016/S2213-2600(20)30247-2 (2020).
Mahony, J. B. et al. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00556-09 (2009).
Wang, D. G., Brewster, J. D., Paul, M. & Tomasula, P. M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules https://doi.org/10.3390/molecules20046048 (2015).
Mori, Y., Hirano, T. & Notomi, T. Sequence specific visual detection of LAMP reactions by addition og cationic polymers. BMC Biotechnol. https://doi.org/10.1186/1472-6750-6-3 (2006).
Phillips, E. A., Moehling, T. J., Bhadra, S., Ellington, A. D. & Linnes, J. C. Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal. Chem. https://doi.org/10.1021/acs.analchem.8b00269 (2018).
De Baar, M. P. et al. One-tube real-time isothermal amplification assay to identify and distinguish human immunodeficiency virus type I subtypes A, B, and C and circulating recombinant forms AE and ag. J. Clin. Microbiol. https://doi.org/10.1128/JCM.39.5.1895-1902.2001 (2001).
Woodford, N. & Ellington, M. J. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. https://doi.org/10.1111/j.1469-0691.2006.01492.x (2007).
Irwin, K. K., Renzette, N., Kowalik, T. F. & Jensen, J. D. Antiviral drug resistance as an adaptive process. Virus Evol. https://doi.org/10.1093/ve/vew014 (2016).
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J. & Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. https://doi.org/10.1007/s00239-004-0046-3 (2005).
Makarova, K. S. et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. https://doi.org/10.1038/nrmicro2577 (2011).
Bolotin, A., Quinquis, B., Sorokin, A. & Dusko Ehrlich, S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology https://doi.org/10.1099/mic.0.28048-0 (2005).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science https://doi.org/10.1126/science.1231143 (2013).
Hilton, I. B. et al. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. https://doi.org/10.1038/nbt.3199 (2015).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science https://doi.org/10.1126/science.aaq0180 (2017).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell https://doi.org/10.1016/j.cell.2013.12.001 (2013).
Sheth, R. U., Yim, S. S., Wu, F. L. & Wang, H. H. Multiplex recording of cellular events over time on CRISPR biological tape. Science https://doi.org/10.1126/science.aao0958 (2017).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. https://doi.org/10.1038/nrmicro3569 (2015).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-019-0299-x (2020).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science 360, 439–444 (2018).
Santiago-Frangos, A. et al. Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell Reports Med. 2, 100319 (2021).
Zhang, Y. et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. https://doi.org/10.1021/acssynbio.6b00215 (2017).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Bao, Y. et al. CUT-LAMP: contamination-free loop-mediated isothermal amplification based on the CRISPR/Cas9 cleavage. ACS Sens. https://doi.org/10.1021/acssensors.0c00034 (2020).
Zhou, W. et al. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. https://doi.org/10.1038/s41467-018-07324-5 (2018).
Compton, J. Nucleic acid sequence-based amplification. Nature https://doi.org/10.1038/350091a0 (1991).
Jiao, C. et al. Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science https://doi.org/10.1126/science.abe7106 (2021).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR–C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Piepenburg, O., Williams, C. H., Stemple, D. L. & Armes, N. A. DNA detection using recombination proteins. PLoS Biol. https://doi.org/10.1371/journal.pbio.0040204 (2006).
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O. & Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. https://doi.org/10.1038/s41596-019-0210-2 (2019).
Yan, W. X. et al. Functionally diverse type V CRISPR–Cas systems. Science https://doi.org/10.1126/science.aav7271 (2019).
Chen, J. S. et al. CRISPR–Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science https://doi.org/10.1126/science.aar6245 (2018).
Li, S.-Y. et al. CRISPR–Cas12a-assisted nucleic acid detection. Cell Discov. 4, 20 (2018).
Li, S. Y. et al. CRISPR–Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. https://doi.org/10.1038/s41422-018-0022-x (2018).
Li, L. et al. HOLMESv2: a CRISPR–Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 8, 2228–2237 (2019).
Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. https://doi.org/10.1093/nar/28.12.e63 (2000).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR–Cas14 enzymes. Science https://doi.org/10.1126/science.aav4294 (2018).
Ramachandran, A. & Santiago, J. G. CRISPR enzyme kinetics for molecular diagnostics. Anal. Chem. https://doi.org/10.1021/acs.analchem.1c00525 (2021).
Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature https://doi.org/10.1038/s41586-020-2196-x (2020).
Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR–Cas13a and mobile phone microscopy. Cell 184, 323–333 (2021).
Hajian, R. et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-019-0371-x (2019).
Bruch, R. et al. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. https://doi.org/10.1002/adma.201905311 (2019).
Niewoehner, O. et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature https://doi.org/10.1038/nature23467 (2017).
Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G. & Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR–Cas systems. Science https://doi.org/10.1126/science.aao0100 (2017).
Moutsiopoulou, A. et al. Bioluminescent protein-inhibitor pair in the design of a molecular aptamer beacon biosensing system. Anal. Chem. https://doi.org/10.1021/acs.analchem.0c00518 (2020).
Storch, G. A. Diagnostic virology. Clin. Infect. Dis. https://doi.org/10.1086/314015 (2000).
Asboe, D. et al. British HIV Association guidelines for the routine investigation and monitoring of adult HIV-1-infected individuals 2011. HIV Med. https://doi.org/10.1111/j.1468-1293.2011.00971.x (2012).
Pawlotsky, J. M. et al. EASL recommendations on treatment of hepatitis C 2018. J. Hepatol. https://doi.org/10.1016/j.jhep.2018.03.026 (2018).
Walker, G. T. et al. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. https://doi.org/10.1093/nar/20.7.1691 (1992).
Vincent, M., Xu, Y. & Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. https://doi.org/10.1038/sj.embor.7400200 (2004).
Van Ness, J., Van Ness, L. K. & Galas, D. J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.0730811100 (2003).
Lillis, L. et al. Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol. Cell. Probes https://doi.org/10.1016/j.mcp.2016.01.009 (2016).
Watzinger, F., Ebner, K. & Lion, T. Detection and monitoring of virus infections by real-time PCR. Mol. Aspects Med. https://doi.org/10.1016/j.mam.2005.12.001 (2006).
Whelan, J. A., Russell, N. B. & Whelan, M. A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods https://doi.org/10.1016/S0022-1759(03)00223-0 (2003).
Schmittgen, T. & Livak, K. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Ding, X. et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR–Cas12a assay. Nat. Commun. 11, 4711 (2020).
Lee, R. A. et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl Acad. Sci. USA 117, 25722–25731 (2020).
Arizti-Sanz, J. et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 11, 5921 (2020).
Joung, J. et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. https://doi.org/10.1056/nejmc2026172 (2020).
Nolan, T., Hands, R. E., Ogunkolade, W. & Bustin, S. A. SPUD: A quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal. Biochem. https://doi.org/10.1016/j.ab.2006.01.051 (2006).
Smith, A. M. et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq368 (2010).
Elnifro, E. M., Ashshi, A. M., Cooper, R. J. & Klapper, P. E. Multiplex PCR: optimization and application in diagnostic virology. Clini. Microbiol. Rev. https://doi.org/10.1128/CMR.13.4.559-570.2000 (2000).
Ackerman, C. M. et al. Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282 (2020).
Gilpatrick, T. et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0407-5 (2020).
Giesselmann, P. et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-019-0293-x (2019).
Gu, W. et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. https://doi.org/10.1186/s13059-016-0904-5 (2016).
Quan, J. et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz418 (2019).
Katzmeier, F. et al. A low-cost fluorescence reader for in vitro transcription and nucleic acid detection with Cas13a. PLoS ONE https://doi.org/10.1371/journal.pone.0220091 (2019).
Yuan, C. et al. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Anal. Chem. https://doi.org/10.1021/acs.analchem.9b05597 (2020).
Hu, M. et al. Single-step, salt-aging-free, and thiol-free freezing construction of AuNP-based bioprobes for advancing CRISPR-based diagnostics. J. Am. Chem. Soc. https://doi.org/10.1021/jacs.0c00217 (2020).
Spoelstra, W. K. et al. CRISPR-based DNA and RNA detection with liquid-liquid phase separation. Biophys. J. 120, 1198–1209 (2021).
Wang, X. et al. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano https://doi.org/10.1021/acsnano.0c00022 (2020).
Dai, Y. et al. Exploring the trans-cleavage activity of CRISPR–Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. Int. Ed. 58, 17399–17405 (2019).
Zhou, T. et al. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Adv. Sci. https://doi.org/10.1002/advs.201903661 (2020).
English, M. A. et al. Programmable CRISPR-responsive smart materials. Science https://doi.org/10.1126/science.aaw5122 (2019).
Gayet, R. V. et al. Creating CRISPR-responsive smart materials for diagnostics and programmable cargo release. Nat. Protoc. https://doi.org/10.1038/s41596-020-0367-8 (2020).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR–Cas13. Science 360, 444–448 (2018).
Ladha, A., Joung, J., Abudayyeh, O. O., Gootenberg, J. S. & Zhang, F. A 5-min RNA preparation method for COVID-19 detection with RT-qPCR. Preprint at medRxiv https://doi.org/10.1101/2020.05.07.20055947 (2020).
Patchsung, M. et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-020-00603-x (2020).
Kaminski, M. M. et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 4, 601–609 (2020).
Qin, P. et al. Rapid and fully microfluidic Ebola virus detection with CRISPR–Cas13a. ACS Sens. https://doi.org/10.1021/acssensors.9b00239 (2019).
Zhang, F., Abudayyeh, O. O., Gootenberg, J. S., Sciences, C. & Mathers, L. A protocol for detection of COVID-19 using CRISPR diagnostics. Bioarchive (2020).
Broughton, J. P. et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0513-4 (2020).
Wu, Y., Liu, S. X., Wang, F. & Zeng, M. S. Room temperature detection of plasma Epstein–Barr virus DNA with CRISPR–Cas13. Clin. Chem. https://doi.org/10.1373/clinchem.2018.299347 (2019).
Ai, J. W. et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 8, 1361–1369 (2019).
Guk, K. et al. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens. Bioelectron. https://doi.org/10.1016/j.bios.2017.04.016 (2017).
Huang, M., Zhou, X., Wang, H. & Xing, D. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. https://doi.org/10.1021/acs.analchem.7b04542 (2018).
Shen, J., Zhou, X. & Shan, Y. et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 11, 267 (2020).
Cunningham, C. H. et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine 68, 103415 (2021).
Shan, Y., Zhou, X., Huang, R. & Xing, D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal. Chem. https://doi.org/10.1021/acs.analchem.9b00073 (2019).
Wang, G., Tian, W., Liu, X., Ren, W. & Liu, C. New CRISPR-derived microRNA sensing mechanism based on Cas12a self-powered and rolling circle transcription-unleashed real-time crRNA recruiting. Anal. Chem. https://doi.org/10.1021/acs.analchem.0c00680 (2020).
SARS-CoV-2 variant classifications and definitions. CDC https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (2021).
Liang, M. et al. A CRISPR–Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 10, 3672 (2019).
Iwasaki, R. S. & Batey, R. T. SPRINT: a Cas13a-based platform for detection of small molecules. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa673 (2020).
Peng, L. et al. CRISPR–Cas12a based aptasensor for sensitive and selective ATP detection. Sens. Actuators B https://doi.org/10.1016/j.snb.2020.128164 (2020).
Xiong, Y. et al. Functional DNA regulated CRISPR–Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J. Am. Chem. Soc. https://doi.org/10.1021/jacs.9b09211 (2020).
Chen, Q., Tian, T., Xiong, E., Wang, P. & Zhou, X. CRISPR/Cas13a signal amplification linked immunosorbent assay for femtomolar protein detection. Anal. Chem. https://doi.org/10.1021/acs.analchem.9b04403 (2020).
Kim, H. K. et al. SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci. Adv. https://doi.org/10.1126/sciadv.aax9249 (2019).
Chuai, G. et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. https://doi.org/10.1186/s13059-018-1459-4 (2018).
Valeri, J. A. et al. Sequence-to-function deep learning frameworks for engineered riboregulators. Nat. Commun. https://doi.org/10.1038/s41467-020-18676-2 (2020).
Angenent-Mari, N. M., Garruss, A. S., Soenksen, L. R., Church, G. & Collins, J. J. A deep learning approach to programmable RNA switches. Nat. Commun. https://doi.org/10.1038/s41467-020-18677-1 (2020).
Shao, N., Han, X., Song, Y., Zhang, P. & Qin, L. CRISPR–Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. https://doi.org/10.1021/acs.analchem.9b02925 (2019).
In vitro companion diagnostic devices. US Food and Drug Administration http://www.fda.gov/regulatory-information/search-fda-guidance-documents/vitro-companion-diagnostic-devices (2019).
Cheng, L., Lopez-Beltran, A., Massari, F., Maclennan, G. T. & Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod. Pathol. https://doi.org/10.1038/modpathol.2017.104 (2018).
Koh, W. et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1405528111 (2014).
Park, S.-m. et al. A mountable toilet system for personalized health monitoring via the analysis of excreta. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-020-0534-9 (2020).
Wang, T., Liu, Y., Sun, H. H., Yin, B. C. & Ye, B. C. An RNA-guided Cas9 nickase-based method for universal isothermal DNA amplification. Angew. Chem. Int. Ed. https://doi.org/10.1002/anie.201901292 (2019).
Teng, F. et al. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 20, 132 (2019).
Acknowledgements
M.M.K. was supported by the Emmy Noether Programme (KA5060/1-1) of the German Research Foundation (DFG). J.J.C. was supported by the Paul G. Allen Frontiers Group and the Wyss Institute. All figures were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing and revising the manuscript, and approved the final version.
Corresponding author
Ethics declarations
Competing interests
J.J.C. is a co-founder and director of Sherlock Biosciences. O.O.A., J.S.G. and F.Z. are co-founders and scientific advisors for Sherlock Biosciences and Pine Trees Health. O.O.A. and J.S.G. are also co-founders and scientific advisors for Moment Biosciences and Tome Biosciences. F.Z. is also a co-founder of Editas Medicine, Beam Therapeutics, Pairwise Plants and Arbor Biotechnologies.
Additional information
Peer review information Nature Biomedical Engineering thanks Kiana Aran, Charles Chiu and Da Xing for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaminski, M.M., Abudayyeh, O.O., Gootenberg, J.S. et al. CRISPR-based diagnostics. Nat Biomed Eng 5, 643–656 (2021). https://doi.org/10.1038/s41551-021-00760-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00760-7
This article is cited by
-
RPA-CRISPR/Cas12a assay for the diagnosis of bovine Anaplasma marginale infection
Scientific Reports (2024)
-
Adeno-associated virus genome quantification with amplification-free CRISPR-Cas12a
Gene Therapy (2024)
-
Massively parallel profiling of RNA-targeting CRISPR-Cas13d
Nature Communications (2024)
-
Topological barrier to Cas12a activation by circular DNA nanostructures facilitates autocatalysis and transforms DNA/RNA sensing
Nature Communications (2024)
-
CRISPR-powered quantitative keyword search engine in DNA data storage
Nature Communications (2024)