Abstract
Cells respond to cytotoxic DNA double-strand breaks (DSBs) by recruiting DNA repair proteins to the damaged site. This recruitment is dependent on ubiquitylation of adjacent chromatin areas by E3 ubiquitin ligases such as RNF8 and RNF168, which are recruited sequentially to the DSBs. However, it is unclear what dictates the sequential order and recruits RNF168 to the DNA lesion. Here, we reveal that L3MBTL2 (lethal(3)malignant brain tumour-like protein 2) is the missing link between RNF8 and RNF168. We found that L3MBTL2 is recruited by MDC1 and subsequently ubiquitylated by RNF8. Ubiquitylated L3MBTL2, in turn, facilitates recruitment of RNF168 to the DNA lesion and promotes DNA DSB repair. These results identify L3MBTL2 as a key target of RNF8 following DNA damage and demonstrates how the DNA damage response pathway is orchestrated by ubiquitin signalling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Price, B. D. & D’Andrea, A. D. Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 (2013).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Goldberg, M. et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421, 952–956 (2003).
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. R. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Wang, B. & Elledge, S. J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl Acad. Sci. USA 104, 20759–20763 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Khan, F. H. et al. Acquired genetic alterations in tumor cells dictate the development of high-risk neuroblastoma and clinical outcomes. BMC Cancer 15, 514 (2015).
Qin, J. et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11, 319–332 (2012).
Stielow, C. et al. SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes. Nucleic Acids Res. 42, 3044–3058 (2014).
Trojer, P. et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 (2011).
Yoo, J. Y. et al. Histone deacetylase 3 is selectively involved in L3MBTL2-mediated transcriptional repression. FEBS Lett. 584, 2225–2230 (2010).
Ogawa, H., Ishiguro, K.-i, Gaubatz, S., Livingston, D. M. & Nakatani, Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 (2002).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Guo, Y. et al. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 37, 2204–2210 (2009).
Acs, K. et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 (2011).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Lou, Z., Minter-Dykhouse, K., Wu, X. & Chen, J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421, 957–961 (2003).
Rodriguez, M., Yu, X., Chen, J. & Songyang, Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J. Biol. Chem. 278, 52914–52918 (2003).
Yu, X., Chini, C. C., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Liu, J. et al. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res. 40, 3898–3912 (2012).
Wu, H.-H., Wu, P.-Y., Huang, K.-F., Kao, Y.-Y. & Tsai, M.-D. Structural delineation of MDC1-FHA domain binding with CHK2-pThr68. Biochemistry 51, 575–577 (2012).
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005).
Lou, Z. et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 (2006).
Bekker-Jensen, S. & Mailand, N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585, 2914–2919 (2011).
Gurvich, N. et al. L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc. Natl Acad. Sci. USA 107, 22552–22557 (2010).
Schwertman, P., Bekker-Jensen, S. & Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016).
Liu, C. et al. RNF168 forms a functional complex with RAD6 during the DNA damage response. J. Cell Sci. 126, 2042–2051 (2013).
Panier, S. et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol. Cell 47, 383–395 (2012).
Penengo, L. et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124, 1183–1195 (2006).
Thorslund, T. et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393 (2015).
Cao, J. & Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2, 26 (2012).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–D520 (2015).
Wang, Z. et al. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 30, 946–959 (2016).
Ramachandran, S. et al. The RNF8/RNF168 ubiquitin ligase cascade facilitates class switch recombination. Proc. Natl Acad. Sci. USA 107, 809–814 (2010).
Peuscher, M. H. & Jacobs, J. J. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat. Cell Biol. 13, 1139–1145 (2011).
Strickfaden, H. et al Poly(ADP-ribosyl)ation-dependent transient chromatin decondensation and histone displacement following laser micro-irradiation.J. Biol. Chem. 291, 1789–1802 (2015).
Deng, Y., Guo, X., Ferguson, D. O. & Chang, S. Multiple roles for MRE11 at uncapped telomeres. Nature 460, 914–918 (2009).
Lou, Z., Chini, C. C. S., Minter-Dykhouse, K. & Chen, J. Mediator of DNA damage checkpoint protein 1 regulates BRCA1 localization and phosphorylation in DNA damage checkpoint control. J. Biol. Chem. 278, 13599–13602 (2003).
Deng, M. et al. Deubiquitination and activation of AMPK by USP10. Mol. Cell 61, 614–624 (2016).
Hamada, M. et al. Ran-dependent docking of importin-β to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 194, 597–612 (2011).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Pei, H. et al. The histone methyltransferase MMSET regulates class switch recombination. J. Immunol. 190, 756–763 (2013).
van Steensel, B., Smogorzewska, A. & de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 (1998).
Acknowledgements
We thank Z. Zhang, M. Jasin, X. Yu, D. Reinberg, N. Dantuma and N. Mailand for providing reagents. Thank you to the members of the Lou lab for helpful discussions and B. Davies for technical assistance. This work was supported, in part, by NIH grants CA130996, CA108961 and CA148940 (to Z.L.) and CA160333 (to Y.D.). S.N. was supported by the Laura J. Siegel Breast Cancer Fellowship Award from the Foundation for Women’s Wellness. K.A. and S.N. thank the Mayo Clinic MSTP for fostering an outstanding environment for physician-scientist training. We thank the Mayo Clinic Cancer Center for the use of the Cytogenetics Core, which provided PNA FISH services. The Mayo Clinic Cancer Center is supported in part by an NCI Cancer Center Support Grant 5P30 CA15083-36.
Author information
Authors and Affiliations
Contributions
S.N., M.D., J.Y., K.L. and B.Q. designed and performed the experiments. K.B.J. and K.A. assisted with the telomere-fusion assay. K.A. and A.A. assisted with confocal microscopy. J.Yu and T.L. provided technical support. H.Z., W.D., Y.D. and J.M.v.D. provided reagents for experiments. S.N. designed experiments, analysed the data and wrote the manuscript with input from the other authors. Z.L. conceived and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
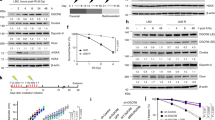
Supplementary Figure 1 The L3MBTL2 antibody used in this study is specific for the protein.
. (a-b) The specificity of the L3MBTL2 antibody was validated by (a) western blot and (b) immunofluorescence. Wild-type or L3MBTL2 knockout U2OS cells were exposed to 2GY irradiation, fixed after an hour, and stained for L3MBTL2 (red). Nucleus was stained with DAPI (blue). Shown are representative images from 3 independent experiments in b. Scale bars, 10 μm. Representative western blots are provided from 3 biologically independent experiments in a. Unprocessed western blots are provided in Supplementary Figure 5
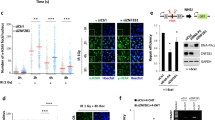
Supplementary Figure 2 L3MBTL2 recruits RNF168 to the double strand break site.
. (a-c) G9A knockdown does not affect RNF168 focus formation in U2OS cells. (d-f) E2F6 knockdown does not affect RNF168 focus formation in U2OS cells. (g-i) PCGF6 knockdown does not affect RNF168 focus formation in U2OS cells. (j-l) L3MBTL1 knockdown does not affect RNF168 focus formation in U2OS cells. (m) MDC1, RNF8 and RNF168 interact with endogenous L3MBTL2 following DNA damage. (n) Knockdown of RNF168 using shRNA does not affect the interaction between RNF8 and L3MBTL2. (o) Knockdown of RNF8 affects the interaction between RNF168 and L3MBTL2. (p) The FHA mutant of MDC1 (R58A) is unable to interact with L3MBTL2 upon DNA damage. The indicated constructs were transfected into MDC1 knockout cells. Cells were exposed to the indicated doses of irradiation. Lysates were collected after an hour and the interaction between RNF168 and L3MBTL2 was assessed. (q) The phosphorylation mutant of L3MBTL2 does not interact with RNF168 upon DNA damage. The indicated constructs were transfected into L3MBTL2 knockout cells. Cells were exposed to the indicated doses of irradiation. Lysates were collected after an hour and the interaction between RNF168 and L3MBTL2 was assessed. (r-t) RNF8 is dispensable for radiation-induced L3MBTL2 foci formation. RNF8 was knocked out using shRNA (GFP-tagged). Cells were exposed to 2GY irradiation, fixed after an hour, and L3MBTL2 foci (red) formation was assessed. Scale bars, 10 μm. Data are represented as the mean ± SEM of n = 3 independent experiments in b, e, h, k and s. Circles depict individual data points. Statistical significance was calculated using 2-way ANOVA. **p = 0.0001 and p=0.001 for control shRNA vs RNF8 shRNA1 and shRNA2 with IR, respectively. Representative western blots are provided from 3 biologically independent experiments in c, f, i, l-q and t. The experiments in a, d, g, j, and r were repeated 3 independent times with similar results. Source data and unprocessed western blots are provided in Supplementary Table 1 and Supplementary Figure 5, respectively
Supplementary Figure 3 RNF8 mediated K63 linked ubiquitylation of L3MBTL2 following DNA damage is critical for the interaction with UDM1 of RNF168 and subsequent histone ubiquitylation.
. (a) Endogenous L3MBTL2 is ubiquitylated by RNF8. Knockdown of RNF8 with shRNA abrogates this DNA damage induced ubiquitylation of L3MBTL2. (b) MDC1 facilitates DNA damage induced L3MBTL2 ubiquitylation. (c) The phosphorylation mutant of L3MBTL2 (S335A) cannot be efficiently ubiquitylated following DNA damage. (d-f) Knockdown of Histone H1 does not affect RNF168 foci formation following DNA damage. (g-i) Comparison of the effect of histone H1 and L3MBTL2 knockdown on various DNA repair proteins. (j-l) HMGB1 does not affect RNF168 foci formation. (m) Histone H1 is mono ubiquitylated following DNA damage. Cells were transfected with the indicated plasmids and exposed to radiation. Lysates were collected after an hour and ubiquitylation of histone H1 was assessed. USP2 was added as indicated and deubiquitylation of histone H1 was assessed to validate that the band observed is indeed histone H1. (n) Histone H1 does not interact with UDM1 following DNA damage. Cell lysates from irradiated cells were incubated with GST-tagged UDM1. Proteins were eluted and blots were probed with the indicated antibodies. (o) H1.x variant of histone H1 does not recognize K63 UIM following DNA damage in MDA-MB-231 cells. (p) Cells were irradiated to induce DNA damage, lysed after 1 hour, and reciprocal endogenous IP was performed with L3MBTL2 and Histone H1 antibodies. No interaction between L3MBTL2 and Histone H1 was observed. Scale bars, 10 μm. Experiments in e and j were repeated 3 independent times with similar results. Data are represented as the mean ± SEM of n = 3 independent experiments in d, i and k. Circles depict individual data points. Statistical significance was calculated using 1-way ANOVA in d and k and 2-way ANOVA in panel i. **p = 0.001 for control vs Histone H1 siRNA with IR in d. **p = 0.000000001 (BRCA1) and p = 0.000000000000007 (53BP1) for control siRNA vs L3MBTL2 siRNA with IR in i. *p = 0.0117 (53BP1) for control siRNA vs Histone H1 siRNA with IR in i. Representative western blots are provided from 3 biologically independent experiments in a-c, f-h, and l-p. Source data and unprocessed western blots are provided in Supplementary Table 1 and Supplementary Figure 5, respectively
Supplementary Figure 4 DNA damage induced RNF8 mediated ubiquitylation of L3MBTL2 is critical for DNA DSB repair.
. (a) Representative images showing RNF168 foci formation in U2OS cells expressing the indicated plasmids (related to Fig. 5e). (b-c) L3MBTL2 knockout or ubiquitylation mutant does not affect protein levels. (d) Representative blot showing the knockdown and knockout efficiency of RNF168 and L3MBTL2 in MDA-MB-231 cells (related to Fig. 5g). (e-f) Loss of L3MBTL2 sensitizes cells to irradiation. (e) Representative blot showing expression levels of exogenous wild-type (WT) or ubiquitylation mutant (K659R) L3MBTL2 in MDA-MB-231 cells. (f) Survival plot of L3MBTL2 knockout MDA-MB-231 cells expressing the indicated plasmids at endogenous levels when exposed to the indicated doses of radiation. (g-h) Loss of L3MBTL2 sensitizes cells to irradiation. (g) Representative blot showing the knockdown and knockout efficiency of RNF168 and L3MBTL2 in U2OS cells. (h) Survival assays of L3MBTL2 knockout U2OS cells treated as indicated and exposed to the indicated doses of irradiation. (i-j) Loss of L3MBTL2 reduces radiation-induced FK2 foci formation. (i) Representative blot showing the knockout efficiency of L3MBTL2. (j) Representative images showing FK2 foci formation in L3MBTL2 control and knockout U2OS cells. (k-l) The MBT domains of L3MBTL2 are important for its role in DNA damage response. (k) Deletion of any of the MBT domains disrupts RNF168 foci formation. (l) Expression level of the various L3MBTL2 constructs utilized in (k). Scale bars, 10 μm. Experiments in a and j were repeated 3 independent times with similar results. Data in f, h, and k are represented as the mean ± SEM of n = 3 independent experiments. Circles depict individual data points. Statistical significance was calculated using 2-way ANOVA in f and h and 1-way ANOVA in panel k. **p = 0.0001 for vector vs all other groups except L3MBTL2 sgRNA + Flag L3MBTL2 WT at each time point in f and h. **p = 0.0000000000001 for WT vs all other MBT deletion groups and p = 0.0004 for WT vs ∆ZN in k. Representative western blots are provided from 3 biologically independent experiments in b-e, g, i and l. Source data and unprocessed western blots are provided in Supplementary Table 1 and Supplementary Figure 5, respectively
Supplementary Figure 5
. Unprocessed images for all blots in this manuscript. Unprocessed images for all blots in this manuscript
Supplementary information
Supplementary Information
Merged Supplementary Figures 1–5 and their legends, and legends for Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Nowsheen, S., Aziz, K., Aziz, A. et al. L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat Cell Biol 20, 455–464 (2018). https://doi.org/10.1038/s41556-018-0071-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0071-x
This article is cited by
-
Protein degradation: expanding the toolbox to restrain cancer drug resistance
Journal of Hematology & Oncology (2023)
-
RNF8 enhances the sensitivity of PD-L1 inhibitor against melanoma through ubiquitination of galectin-3 in stroma
Cell Death Discovery (2023)
-
Dynamic recruitment of UFM1-specific peptidase 2 to the DNA double-strand breaks regulated by WIP1
Genome Instability & Disease (2022)
-
METTL16 antagonizes MRE11-mediated DNA end resection and confers synthetic lethality to PARP inhibition in pancreatic ductal adenocarcinoma
Nature Cancer (2022)
-
Reciprocal regulation of RIG-I and XRCC4 connects DNA repair with RIG-I immune signaling
Nature Communications (2021)