Abstract
Candidate phyla radiation (CPR) bacteria and DPANN (an acronym of the names of the first included phyla) archaea are massive radiations of organisms that are widely distributed across Earth’s environments, yet we know little about them. Initial indications are that they are consistently distinct from essentially all other bacteria and archaea owing to their small cell and genome sizes, limited metabolic capacities and often episymbiotic associations with other bacteria and archaea. In this Analysis, we investigate their biology and variations in metabolic capacities by analysis of approximately 1,000 genomes reconstructed from several metagenomics-based studies. We find that they are not monolithic in terms of metabolism but rather harbour a diversity of capacities consistent with a range of lifestyles and degrees of dependence on other organisms. Notably, however, certain CPR and DPANN groups seem to have exceedingly minimal biosynthetic capacities, whereas others could potentially be free living. Understanding of these microorganisms is important from the perspective of evolutionary studies and because their interactions with other organisms are likely to shape natural microbiome function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


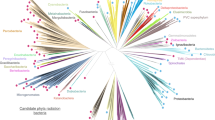
Adapted from ref.61, Annual Reviews.





Similar content being viewed by others
Change history
04 October 2018
In the original online version of this manuscript, the ORCID links for Cindy J. Castelle, Karthik Anantharaman, Alexander J. Probst and Jillian F. Banfield were omitted. These have now been added.
References
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Castelle, C. J. et al. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701 (2015).
Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015).
Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016).
Anantharaman, K. et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016).
Wrighton, K. C. et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665 (2012).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Huber, H. et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417, 63–67 (2002).
Baker, B. J. et al. Lineages of acidophilic archaea revealed by community genomic analysis. Science 314, 1933–1935 (2006).
Baker, B. J. et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl Acad. Sci. USA 107, 8806–8811 (2010).
Probst, A. J. et al. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat. Microbiol. 3, 328–336 (2018).
Probst, A. J. et al. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ. Microbiol. 19, 459–474 (2017).
Williams, T. A. et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl Acad. Sci. USA 114, E4602–E4611 (2017).
Adam, P. S., Borrel, G., Brochier-Armanet, C. & Gribaldo, S. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11, 2407–2425 (2017).
Aouad, M. et al. Extreme halophilic archaea derive from two distinct methanogen Class II lineages. Mol. Phylogenet. Evol. 127, 46–54 (2018).
Petitjean, C., Deschamps, P., López-García, P. & Moreira, D. Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biol. Evol. 7, 191–204 (2014).
Hua, Z.-S. et al. Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J. 9, 1280–1294 (2015).
Suzuki, S. et al. Microbial diversity in The Cedars, an ultrabasic, ultrareducing, and low salinity serpentinizing ecosystem. Proc. Natl Acad. Sci. USA 110, 15336–15341 (2013).
Narasingarao, P. et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6, 81–93 (2012).
Andrade, K. et al. Metagenomic and lipid analyses reveal a diel cycle in a hypersaline microbial ecosystem. ISME J. 9, 2697–2711 (2015).
Ortiz-Alvarez, R. & Casamayor, E. O. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ. Microbiol. Rep. 8, 210–217 (2016).
Linz, A. M. et al. Bacterial community composition and dynamics spanning five years in freshwater bog lakes. mSphere 2, e00169–00117 (2017).
Merkley, E. D. et al. Changes in protein expression across laboratory and field experiments in Geobacter bemidjiensis. J. Proteome Res. 14, 1361–1375 (2015).
Ludington, W. B. et al. Assessing biosynthetic potential of agricultural groundwater through metagenomic sequencing: a diverse anammox community dominates nitrate-rich groundwater. PLOS One 12, e0174930 (2017).
Lin, X., Kennedy, D., Fredrickson, J., Bjornstad, B. & Konopka, A. Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ. Microbiol. 14, 414–425 (2012).
Tully, B. J., Graham, E. D. & Heidelberg, J. F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018).
Wright, J. J., Konwar, K. M. & Hallam, S. J. Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394 (2012).
Li, M. et al. Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat. Commun. 6, 8933 (2015).
Dombrowski, N., Seitz, K. W., Teske, A. P. & Baker, B. J. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5, 106 (2017).
Schauer, C., Thompson, C. L. & Brune, A. The bacterial community in the gut of the Cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl. Environ. Microbiol. 78, 2758–2767 (2012).
Biedermann, L. et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLOS One 8, e59260 (2013).
Camanocha, A. & Dewhirst, F. E. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J. Oral Microbiol. 6, 25468 (2014).
Thomas, T. et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 11870 (2016).
Koskinen, K. et al. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. MBio 8, e00824–00817 (2017).
Bruno, A. et al. Exploring the under-investigated “microbial dark matter” of drinking water treatment plants. Sci. Rep. 7, 44350 (2017).
Bautista-de los Santos, Q. M. et al. Emerging investigators series: microbial communities in full-scale drinking water distribution systems — a meta-analysis. Environ. Sci. Water Res. Technol. 2, 631–644 (2016).
Pinto, A. J., Schroeder, J., Lunn, M., Sloan, W. & Raskin, L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. MBio 5, e01135–01114 (2014).
Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
He, X. et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl Acad. Sci. USA 112, 244–249 (2015).
Ling, Z. et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 80, 2546–2554 (2014).
Kuehbacher, T. et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57, 1569–1576 (2008).
Kowarsky, M. et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc. Natl Acad. Sci. USA 114, 9623–9628 (2017).
Dudek, N. K. et al. Novel microbial diversity and functional potential in the marine mammal oral microbiome. Curr. Biol. 27, 3752–3762 (2017).
Golyshina, O. V. et al. ‘ARMAN’ archaea depend on association with euryarchaeal host in culture and in situ. Nat. Commun. 8, 60 (2017).
Youssef, N. H. et al. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites’. ISME J. 9, 447–460 (2014).
Nelson, W. C. & Stegen, J. C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6, 713 (2015).
Kantor, R. S. et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4, e00708–e00713 (2013).
Wrighton, K. C. et al. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 8, 1452–1463 (2014).
Campbell, J. H. et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc. Natl Acad. Sci. USA 110, 5540–5545 (2013).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Erickson, H. P. & Osawa, M. Cell division without FtsZ — a variety of redundant mechanisms. Mol. Microbiol. 78, 267–270 (2010).
Luef, B. et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 6, 6372 (2015).
Huber, H., Hohn, M. J., Rachel, R. & Fuchs, T. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. 417, 63–67 (2002).
Junglas, B. et al. Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch. Microbiol. 190, 395–408 (2008).
Burghardt, T. et al. The interaction of Nanoarchaeum equitans with Ignicoccus hospitalis: proteins in the contact site between two cells. Biochem. Soc. Trans. 37, 127–132 (2009).
Wurch, L. et al. ARTICLE Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment. Nat. Commun. 7, 12115 (2016).
Comolli, L. R. & Banfield, J. F. Inter-species interconnections in acid mine drainage microbial communities. Front. Microbiol. 5, 367 (2014).
Soro, V. et al. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl. Environ. Microbiol. 80, 6480–6489 (2014).
Brown, C. T., Olm, M. R., Thomas, B. C. & Banfield, J. F. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016).
Moran, N. A. & Wernegreen, J. J. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15, 321–326 (2000).
Moran, N. A. & Bennett, G. M. The tiniest tiny genomes. Annu. Rev. Microbiol. 68, 195–215 (2014).
Lenhart, J. S. et al. RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J. Bacteriol. 196, 2851–2860 (2014).
Burstein, D. et al. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 7, 10613 (2016).
Chen, I. & Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 (2004).
Brasen, C., Esser, D., Rauch, B. & Siebers, B. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175 (2014).
Silva, P. J. et al. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551 (2000).
Wrighton, K. C. et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 10, 2702–2714 (2016).
Aono, R. et al. Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J. Bacteriol. 194, 6847–6855 (2012).
Hernsdorf, A. W. et al. Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J. 11, 1915–1929 (2017).
Anantharaman, K. et al. Analysis of five complete genome sequences for members of the class Peribacteria in the recently recognized Peregrinibacteria bacterial phylum. PeerJ 4, e1607 (2016).
Castelle, C. J., Brown, C. T., Thomas, B. C., Williams, K. H. & Banfield, J. F. Unusual respiratory capacity and nitrogen metabolism in a Parcubacterium (OD1) of the Candidate Phyla Radiation. Sci. Rep. 7, 40101 (2017).
Danczak, R. E. et al. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5, 112 (2017).
León-Zayas, R. et al. The metabolic potential of the single cell genomes obtained from the Challenger Deep, Mariana Trench within the Candidate Superphylum Parcubacteria (OD1). Environ. Microbiol. 19, 2769–2784 (2017).
Coursolle, D. & Gralnick, J. A. Reconstruction of extracellular respiratory pathways for iron(III) reduction in Shewanella Oneidensis strain MR-1. Front. Microbiol. 3, 56 (2012).
Liu, J. et al. Identification and characterization of MtoA: a Decaheme c-Type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3, 37 (2012).
Jiao, Y. & Newman, D. K. The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773 (2007).
Yan, Z, Wang, M. & Ferry, J. G. A ferredoxin-and F 420 H 2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains Bacteria and Archaea. 8, 2285–2301 (2017).
Yan, Z. & Ferry, J. G. Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9, 1322 (2018).
Castelle, C. J. & Banfield, J. F. Major new microbial groups expand diversity and alter our understanding of the Tree of Life. Cell 172, 1181–1197 (2018).
Gross, C. A. et al. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 63, 141–155 (1998).
Paget, M. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5, 1245–1265 (2015).
Merrick, M. J. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10, 903–909 (1993).
Mukai, T., Reynolds, N., Crnkovic´, A. & Söll, D. Bioinformatic analysis reveals archaeal tRNATyr and tRNATrp identities in bacteria. Life 7, 8 (2017).
Armengod, M.-E. et al. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 94, 1510–1520 (2012).
Selvadurai, K., Wang, P., Seimetz, J. & Huang, R. H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 10, 810–812 (2014).
Krogan, N. J. & Greenblatt, J. F. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8203–8212 (2001).
Glatt, S. et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 23, 794–802 (2016).
Guo, H., Arambula, D., Ghosh, P. & Miller, J. F. Diversity-generating retroelements in phage and bacterial genomes. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MDNA3-0029-2014 (2014).
Paul, B. G. et al. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 6, 6585 (2015).
Paul, B. G. et al. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat. Microbiol. 2, 17045 (2017).
Salman, V., Amann, R., Shub, D. A. & Schulz-Vogt, H. N. Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria. Proc. Natl Acad. Sci. USA 109, 4203–4208 (2012).
Baker, B. J., Hugenholtz, P., Dawson, S. C. & Banfield, J. F. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 69, 5512–5518 (2003).
Jay, Z. J. & Inskeep, W. P. The distribution, diversity, and importance of 16S rRNA gene introns in the order Thermoproteales. Biol. Direct 10, 35 (2015).
Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A. & Eddy, S. R. Rfam: an RNA family database. Nucleic Acids Res. 31, 439–441 (2003).
Tanaka, N., Meineke, B. & Shuman, S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 286, 30253–30257 (2011).
Shah, N. H. & Muir, T. W. Inteins: nature’s gift to protein chemists. Chem. Sci. 5, 446–461 (2014).
Gong, J., Qing, Y., Guo, X. & Warren, A. ‘Candidatus Sonnebornia yantaiensis’, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora. Oligohymenophorea). Syst. Appl. Microbiol. 37, 35–41 (2014).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Burstein, D. et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241 (2016).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Acknowledgements
Support was provided by grants from the Lawrence Berkeley National Laboratory’s Genomes-to-Watershed Scientific Focus Area. The US Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research funded the work under contract DE-AC02-05CH11231, the DOE carbon cycling programme DOE-SC10010566, the Sloan Foundation Deep Life (grant number G-2016-20166041), the Innovative Genomics Institute at the University of California, Berkeley and the Chan Zuckerberg Biohub. Sequencing was conducted by the DOE Joint Genome Institute, a DOE Office of Science User Facility, supported under contract number DE-AC02-05CH11231.
Reviewer information
Nature Reviews Microbiology thanks B. Baker, P. López-García, M. Strous and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.J.C., J.F.B. and C.T.B. conducted new data analysis. C.T.B., K.A. and R.H.H. contributed to the discussion of content. C.J.C. and J.F.B. wrote the article, and J.F.B, C.J.C., C.T.B., K.A., A.J.P. and R.H.H. reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
ggKbase database: https://ggkbase.berkeley.edu/genome_summaries/1437-DPANN_CPR_overall_analyses_NRM_updated
Supplementary information
Glossary
- Candidate phyla
-
A phylum that is defined on the basis of sequence information and lacks any isolated representative.
- Monophyletic
-
A group of organisms that arose from a common ancestor.
- Episymbionts
-
Symbionts that are attached to the surface of another cell.
Rights and permissions
About this article
Cite this article
Castelle, C.J., Brown, C.T., Anantharaman, K. et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16, 629–645 (2018). https://doi.org/10.1038/s41579-018-0076-2
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0076-2
This article is cited by
-
Global abundance patterns, diversity, and ecology of Patescibacteria in wastewater treatment plants
Microbiome (2024)
-
Autotrophic biofilms sustained by deeply sourced groundwater host diverse bacteria implicated in sulfur and hydrogen metabolism
Microbiome (2024)
-
Expanded phylogeny of extremely halophilic archaea shows multiple independent adaptations to hypersaline environments
Nature Microbiology (2024)
-
Functional and evolutionary significance of unknown genes from uncultivated taxa
Nature (2024)
-
Variable impact of geochemical gradients on the functional potential of bacteria, archaea, and phages from the permanently stratified Lac Pavin
Microbiome (2023)