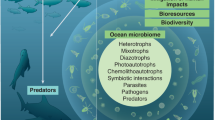
Abstract
A planetary-scale understanding of the ocean ecosystem, particularly in light of climate change, is crucial. Here, we review the work of Tara Oceans, an international, multidisciplinary project to assess the complexity of ocean life across comprehensive taxonomic and spatial scales. Using a modified sailing boat, the team sampled plankton at 210 globally distributed sites at depths down to 1,000 m. We describe publicly available resources of molecular, morphological and environmental data, and discuss how an ecosystems biology approach has expanded our understanding of plankton diversity and ecology in the ocean as a planetary, interconnected ecosystem. These efforts illustrate how global-scale concepts and data can help to integrate biological complexity into models and serve as a baseline for assessing ecosystem changes and the future habitability of our planet in the Anthropocene epoch.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998).
Guidi, L. et al. A new look at ocean carbon remineralization for estimating deepwater sequestration. Global Biogeochem. Cycles 29, 1044–1059 (2015).
Henson, S. A., Sanders, R. & Madsen, E. Global patterns in efficiency of particulate organic carbon export and transfer to the deep ocean. Global Biogeochem. Cycles 26, GB1028 (2012).
Kwon, E. Y., Primeau, F. & Sarmiento, J. L. The impact of remineralization depth on the air-sea carbon balance. Nat. Geosci. 2, 630–635 (2009).
Azam, F. et al. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263 (1983).
Raes, J. & Bork, P. Molecular eco-systems biology: towards an understanding of community function. Nat. Rev. Microbiol. 6, 693–699 (2008).
Karsenti, E. et al. A holistic approach to marine eco-systems biology. PLoS Biol. 9, e1001177 (2011).
Rusch, D. B. et al. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77 (2007).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004). This study applies high-throughput DNA sequencing to produce a large data set of microbial community genome fragments from surface seawaters of the Sargasso Sea and identifies more than 1.2 million previously unknown genes, illustrating the diversity of ocean microbial life.
Yooseph, S. et al. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 5, e16 (2007).
Pesant, S. et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2, 150023 (2015).
Biller, S. J. et al. Marine microbial metagenomes sampled across space and time. Sci. Data 5, 180176 (2018).
Duarte, C. M. Seafaring in the 21st century: the Malaspina 2010 Circumnavigation Expedition. Limnol. Oceanogr. Bull. 24, 11–14 (2015).
Karl, D. M. & Church, M. J. Microbial oceanography and the Hawaii ocean time-series programme. Nat. Rev. Microbiol. 12, 699–713 (2014).
Kopf, A. et al. The ocean sampling day consortium. Gigascience 4, 27 (2015).
Amaral-Zettler, L. et al. in Life in the World’s Oceans (ed. McIntyre, A. D.) 221–245 (Wiley, 2010).
Longhurst, A. Seasonal cycles of pelagic production and consumption. Prog. Oceanogr. 36, 77–167 (1995).
Sunagawa, S., Karsenti, E., Bowler, C. & Bork, P. Computational eco-systems biology in Tara Oceans: translating data into knowledge. Mol. Syst. Biol. 11, 809 (2015).
Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999).
Suttle, C. A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Wommack, K. E. & Colwell, R. R. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114 (2000).
Brum, J. R. & Sullivan, M. B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 (2015).
Brum, J. R. et al. Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). This article describes the first of the Tara Oceans efforts to investigate the diversity and structure of double-stranded DNA viral communities in the oceans, supporting a model of passive global transport by ocean currents and selection by local environmental conditions.
Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123.e14 (2019).
Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016).
Duhaime, M. B. et al. Comparative omics and trait analyses of marine pseudoalteromonas phages advance the phage OTU concept. Front. Microbiol. 8, 1241 (2017).
Martinez-Hernandez, F. et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 8, 15892 (2017).
Warwick-Dugdale, J. et al. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ 7, e6800 (2019).
Nishimura, Y. et al. Environmental viral genomes shed new light on virus-host interactions in the ocean. mSphere 2, e00359-16 (2017).
Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379–2380 (2017).
Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019).
Bolduc, B. et al. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and bacteria. PeerJ 5, e3243 (2017).
Roux, S. et al. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 37, 29–37 (2019).
Simmonds, P. et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161–168 (2017).
Baas-Becking, L. G. M. Geobiologie of Inleiding tot de Milieukunde (Van Stockum & Zoon, 1934).
Ibarbalz, F. M. et al. Global trends in marine plankton diversity across kingdoms of life. Cell 179, 1084–1097.e21 (2019).
Jia, Y., Shan, J., Millard, A., Clokie, M. R. & Mann, N. H. Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol. Lett. 310, 120–126 (2010).
Ribalet, F. et al. Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc. Natl Acad. Sci. USA 112, 8008–8012 (2015).
Yoshida, T. et al. Locality and diel cycling of viral production revealed by a 24 h time course cross-omics analysis in a coastal region of Japan. ISME J. 12, 1287–1295 (2018).
Fridman, S. et al. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2, 1350–1357 (2017).
Mann, N. H., Cook, A., Millard, A., Bailey, S. & Clokie, M. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424, 741 (2003).
Sullivan, M. B. et al. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4, e234 (2006).
Hurwitz, B. L., Hallam, S. J. & Sullivan, M. B. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 14, R123 (2013).
Howard-Varona, C. et al. Regulation of infection efficiency in a globally abundant marine Bacteriodetes virus. ISME J. 11, 284–295 (2017).
Guidi, L. et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). This study integrates Tara Oceans data across organismal size classes from epipelagic depths, revealing that unexpected taxa can predict the downward export of carbon by biological processes in subtropical, nutrient-depleted oceans.
Howard-Varona, C. et al. Phage-specific metabolic reprogramming of virocells. ISME J. 14, 881–895 (2020).
Wilhelm, S. W. & Suttle, C. A. Viruses and nutrient cycles in the sea - viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788 (1999).
Hurwitz, B. L., Brum, J. R. & Sullivan, M. B. Depth-stratified functional and taxonomic niche specialization in the ‘core’ and ‘flexible’ Pacific Ocean virome. ISME J. 9, 472–484 (2015).
Carradec, Q. et al. A global ocean atlas of eukaryotic genes. Nat. Commun. 9, 373 (2018).
Hingamp, P. et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 7, 1678–1695 (2013).
Lescot, M. et al. Reverse transcriptase genes are highly abundant and transcriptionally active in marine plankton assemblages. ISME J. 10, 1134–1146 (2016).
Villar, E. et al. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science 348, 1261447 (2015).
Li, Y. et al. The earth is small for “Leviathans”: long distance dispersal of giant viruses across aquatic environments. Microbes Environ. 34, 334–339 (2019).
Mihara, T. et al. Taxon richness of “Megaviridae” exceeds those of bacteria and archaea in the ocean. Microbes Environ. 33, 162–171 (2018).
von Dassow, P. et al. Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 9, 1365–1377 (2015).
Clerissi, C. et al. Deep sequencing of amplified Prasinovirus and host green algal genes from an Indian Ocean transect reveals interacting trophic dependencies and new genotypes. Environ. Microbiol. Rep. 7, 979–989 (2015).
Clerissi, C. et al. Unveiling of the diversity of prasinoviruses (Phycodnaviridae) in marine samples by using high-throughput sequencing analyses of PCR-amplified DNA polymerase and major capsid protein genes. Appl. Environ. Microbiol. 80, 3150–3160 (2014).
Clerissi, C. et al. Prasinovirus distribution in the northwest Mediterranean Sea is affected by the environment and particularly by phosphate availability. Virology 466–467, 146–157 (2014).
Li, Y. et al. Degenerate PCR primers to reveal the diversity of giant viruses in coastal waters. Viruses 10 (2018).
Blanc-Mathieu, R. et al. Viruses of the eukaryotic plankton are predicted to increase carbon export efficiency in the global sunlit ocean. Preprint at bioRxiv https://doi.org/10.1101/710228 (2019).
Bolduc, B., Youens-Clark, K., Roux, S., Hurwitz, B. L. & Sullivan, M. B. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 11, 7–14 (2017).
Mihara, T. et al. Linking virus genomes with host taxonomy. Viruses 8, 66 (2016).
Steward, G. F. et al. Are we missing half of the viruses in the ocean? ISME J. 7, 672–679 (2013).
Salazar, G. et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179, 1068–1083.e21 (2019).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). This study catalogues 40 million ocean microbial genes and shows temperature to be a main driver of open-ocean microbial community composition in the epipelagic zone at a global scale.
Kultima, J. R. et al. MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS One 7, e47656 (2012).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
DeLong, E. F. et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503 (2006).
Giovannoni, S. J. & Stingl, U. Molecular diversity and ecology of microbial plankton. Nature 437, 343–348 (2005).
Fuhrman, J. A. et al. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl Acad. Sci. USA 103, 13104–13109 (2006).
Milanese, A. et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 10, 1014 (2019).
Farrant, G. K. et al. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc. Natl Acad. Sci. USA 113, E3365–E3374 (2016).
Grebert, T. et al. Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc. Natl Acad. Sci. USA 115, E2010–E2019 (2018).
Yelton, A. P. et al. Global genetic capacity for mixotrophy in marine picocyanobacteria. ISME J. 10, 2946–2957 (2016).
Royo-Llonch, M. et al. Exploring microdiversity in novel Kordia sp. (Bacteroidetes) with proteorhodopsin from the tropical Indian Ocean via single amplified genomes. Front. Microbiol. 8, 1317 (2017).
Royo-Llonch, M., Sánchez, P., González, J. M., Pedrós-Alió, C. & Acinas, S. G. Ecological and functional capabilities of an uncultured Kordia sp. Syst. Appl. Microbiol. 43, 126045 (2020).
Cabello, A. M. et al. Global distribution and vertical patterns of a prymnesiophyte-cyanobacteria obligate symbiosis. ISME J. 10, 693–706 (2016).
Cornejo-Castillo, F. M. et al. Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nat. Commun. 7, 11071 (2016).
Cornejo-Castillo, F. M. et al. UCYN-A3, a newly characterized open ocean sublineage of the symbiotic N2 -fixing cyanobacterium Candidatus Atelocyanobacterium thalassa. Environ. Microbiol. 21, 111–124 (2019).
Delmont, T. O. et al. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813 (2018).
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018). This study exemplifies the use of Tara Oceans data to formulate new hypotheses by reconstructing genomes that support a mitochondrial origin before the divergence of all Alphaproteobacteria sampled to date.
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Tully, B. J., Graham, E. D. & Heidelberg, J. F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018).
Pushkarev, A. et al. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595–599 (2018).
Oppermann, J. et al. MerMAIDs: a family of metagenomically discovered marine anion-conducting and intensely desensitizing channelrhodopsins. Nat. Commun. 10, 3315 (2019).
Louca, S., Parfrey, L. W. & Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277 (2016).
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc. Natl Acad. Sci. USA 115, 6506–6511 (2018).
Caron, D. A., Countway, P. D., Jones, A. C., Kim, D. Y. & Schnetzer, A. Marine protistan diversity. Ann. Rev. Mar. Sci. 4, 467–493 (2012).
Colin, S. et al. Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes. eLife 6, e26066 (2017).
Decelle, J. et al. PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 15, 1435–1445 (2015).
Guillou, L. et al. The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013).
Seeleuthner, Y. et al. Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat. Commun. 9, 310 (2018).
Sieracki, M. E. et al. Single cell genomics yields a wide diversity of small planktonic protists across major ocean ecosystems. Sci. Rep. 9, 6025 (2019).
de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). This study surveys the eukaryotic diversity of ocean plankton from the smallest protists to millimetre-sized animals by 18S ribosomal RNA gene amplicon sequencing, revealing 150,000 taxonomic groups dominated by protistan parasites and symbiotic hosts.
Flegontova, O. et al. Extreme diversity of diplonemid eukaryotes in the ocean. Curr. Biol. 26, 3060–3065 (2016).
Decelle, J. et al. Worldwide occurrence and activity of the reef-building coral symbiont Symbiodinium in the open ocean. Curr. Biol. 28, 3625–3633 e3623 (2018).
Lima-Mendez, G. et al. Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015). This study evaluates the effect of abiotic and biotic factors on organismal interactions among bacteria, archaea, eukaryotes and viruses, emphasizing the role of grazing, pathogenicity and parasitism as predictors of plankton community structure.
Vincent, F. J. et al. The epibiotic life of the cosmopolitan diatom Fragilariopsis doliolus on heterotrophic ciliates in the open ocean. ISME J. 12, 1094–1108 (2018).
Malviya, S. et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl Acad. Sci. USA 113, E1516–E1525 (2016).
Le Bescot, N. et al. Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ. Microbiol. 18, 609–626 (2016).
Lopes Dos Santos, A. et al. Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters. ISME J. 11, 512–528 (2017).
Gimmler, A., Korn, R., de Vargas, C., Audic, S. & Stoeck, T. The Tara Oceans voyage reveals global diversity and distribution patterns of marine planktonic ciliates. Sci. Rep. 6, 33555 (2016).
Beaugrand, G., Luczak, C., Goberville, E. & Kirby, R. R. Marine biodiversity and the chessboard of life. PLoS One 13, e0194006 (2018).
Biard, T. et al. Biogeography and diversity of Collodaria (Radiolaria) in the global ocean. ISME J. 11, 1331–1344 (2017).
Del Campo, J. et al. Assessing the diversity and distribution of apicomplexans in host and free-living environments using high-throughput amplicon data and a phylogenetically informed reference framework. Front. Microbiol. 10, 2373 (2019).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Foster, Z. S., Sharpton, T. J. & Grunwald, N. J. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 13, e1005404 (2017).
Pierella Karlusich, J. J., Ibarbalz, F. M. & Bowler, C. Phytoplankton in the Tara Ocean. Annu. Rev. Mar. Sci. 12, 233–265 (2020).
Leblanc, K. et al. Nanoplanktonic diatoms are globally overlooked but play a role in spring blooms and carbon export. Nat. Commun. 9, 953 (2018).
Treguer, P. et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37 (2018).
Rabosky, D. L. & Sorhannus, U. Diversity dynamics of marine planktonic diatoms across the Cenozoic. Nature 457, 183–186 (2009).
Azaele, S., Pigolotti, S., Banavar, J. R. & Maritan, A. Dynamical evolution of ecosystems. Nature 444, 926–928 (2006).
Ferriere, R. & Cazelles, B. Universal power laws govern intermittent rarity in communities of interacting species. Ecology 80, 1505–1521 (1999).
Gawryluk, R. M. R. et al. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 26, 3053–3059 (2016).
Mordret, S. et al. The symbiotic life of Symbiodinium in the open ocean within a new species of calcifying ciliate (Tiarina sp.). ISME J. 10, 1424–1436 (2016).
Biard, T. et al. In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532, 504–507 (2016).
Vannier, T. et al. Survey of the green picoalga Bathycoccus genomes in the global ocean. Sci. Rep. 6, 37900 (2016).
Steinberg, D. K. & Landry, M. R. Zooplankton and the ocean carbon cycle. Ann. Rev. Mar. Sci. 9, 413–444 (2017).
Roullier, F. et al. Particle size distribution and estimated carbon flux across the Arabian Sea oxygen minimum zone. Biogeosciences 11, 4541–4557 (2014).
Corse, E. et al. Phylogenetic analysis of Thecosomata Blainville, 1824 (holoplanktonic opisthobranchia) using morphological and molecular data. PLoS One 8, e59439 (2013).
Gasmi, S. et al. Evolutionary history of Chaetognatha inferred from molecular and morphological data: a case study for body plan simplification. Front. Zool. 11, 84 (2014).
Madoui, M. A. et al. New insights into global biogeography, population structure and natural selection from the genome of the epipelagic copepod Oithona. Mol. Ecol. 26, 4467–4482 (2017).
Arif, M. et al. Discovering millions of plankton genomic markers from the Atlantic Ocean and the Mediterranean Sea. Mol. Ecol. Resour. 19, 526–535 (2019).
Caputi, L. et al. Community-level responses to iron availability in open ocean plankton ecosystems. Global Biogeochem. Cycles 33, 391–419 (2019).
Busseni, G. et al. Meta-omics reveals genetic flexibility of diatom nitrogen transporters in response to environmental changes. Mol. Biol. Evol. 36, 2522–2535 (2019).
D’Alelio, D. et al. Modelling the complexity of plankton communities exploiting omics potential: From present challenges to an integrative pipeline. Curr. Opin. Syst. Biol. 13, 68–74 (2019).
Whittaker, R. H. Evolution and measurement of species diversity. Taxon 21, 213–251 (1972).
Fuhrman, J. A. et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl Acad. Sci. USA 105, 7774–7778 (2008).
Raes, E. J. et al. Oceanographic boundaries constrain microbial diversity gradients in the South Pacific Ocean. Proc. Natl Acad. Sci. USA 115, E8266–E8275 (2018).
Capotondi, A. et al. Observational needs supporting marine ecosystems modeling and forecasting: from the global ocean to regional and coastal systems. Front. Mar. Sci. 6, 623 (2019).
Lombard, F. et al. Globally consistent quantitative observations of planktonic ecosystems. Front. Mar. Sci. 6, 196 (2019).
Ten Hoopen, P. et al. Marine microbial biodiversity, bioinformatics and biotechnology (M2B3) data reporting and service standards. Stand. Genomic Sci. 10, 20 (2015).
Gorsky, G. et al. Expanding Tara Oceans protocols for underway, ecosystemic sampling of the ocean-atmosphere interface during Tara Pacific expedition (2016–2018). Front. Mar. Sci. 6, 750 (2019).
Planes, S. et al. The Tara Pacific expedition — a pan-ecosystemic approach of the “-omics” complexity of coral reef holobionts across the Pacific Ocean. PLoS Biol. 17, e3000483 (2019).
Bolhuis, H. et al. Atlantic Ocean Research Alliance — marine microbiome roadmap (AORA, 2020).
Cram, J. A. et al. Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J. 9, 563–580 (2015).
D’Alcala, M. R. et al. Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Sci. Mar. 68, 65–83 (2004).
Gilbert, J. A. et al. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS One 5, e15545 (2010).
Romagnan, J. B. et al. Comprehensive model of annual plankton succession based on the whole-plankton time series approach. PLoS One 10, e0119219 (2015).
Gasol, J. M. et al. ICES phytoplankton and microbial plankton status report 2009/2010 (eds O’Brien, T. D., Li, W. K. W. & Morán, X. A. G.) 138–141 (ICES, 2012).
Martin-Platero, A. M. et al. High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat. Commun. 9, 266 (2018).
Laber, C. P. et al. Coccolithovirus facilitation of carbon export in the North Atlantic. Nat. Microbiol. 3, 537–547 (2018).
Marx, V. When microbiologists plunge into the ocean. Nat. Methods 17, 133–136 (2020).
Buttigieg, P. L. et al. Marine microbes in 4D-using time series observation to assess the dynamics of the ocean microbiome and its links to ocean health. Curr. Opin. Microbiol. 43, 169–185 (2018).
Shneider, A. M. Four stages of a scientific discipline; four types of scientist. Trends Biochem. Sci. 34, 217–223 (2009).
Karl, D. M. A sea of change: biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems 2, 181–214 (1999).
Cavicchioli, R. et al. Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586 (2019). This review article provides a consensus statement, the ‘microbiologists’ warning to humanity’, documenting how microorganisms will affect and will be affected by climate change.
Bork, P. et al. Tara Oceans studies plankton at planetary scale. Introduction. Science 348, 873 (2015).
Logares, R. et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ. Microbiol. 16, 2659–2671 (2014).
Nakayama, T. et al. Single-cell genomics unveiled a cryptic cyanobacterial lineage with a worldwide distribution hidden by a dinoflagellate host. Proc. Natl Acad. Sci. USA 116, 15973–15978 (2019).
Probert, I. et al. Brandtodinium gen. nov. and B. nutricula comb. Nov. (Dinophyceae), a dinoflagellate commonly found in symbiosis with polycystine radiolarians. J. Phycol. 50, 388–399 (2014).
Decelle, J., Colin, S. & Foster, R. A. in Marine Protists: Diversity and Dynamics (eds Ohtsuka, S. et al.) 465–500 (Springer, 2015).
Acknowledgements
Tara Oceans (which includes the Tara Oceans and Tara Oceans Polar Circle expeditions) would not exist without the leadership of the Tara Ocean Foundation and the continuous support of 23 institutes (https://oceans.taraexpeditions.org/). The authors further thank the commitment of the following sponsors: the French CNRS (in particular Groupement de Recherche GDR3280 and the Research Federation for the Study of Global Ocean Systems Ecology and Evolution FR2022/Tara GOSEE), the French Facility for Global Environment (FFEM), the European Molecular Biology Laboratory, Genoscope/CEA, the French Ministry of Research and the French Government Investissements d’Avenir programmes OCEANOMICS (ANR-11-BTBR-0008), FRANCE GENOMIQUE (ANR-10-INBS-09-08) and MEMO LIFE (ANR-10-LABX-54), the PSL research university (ANR-11-IDEX-0001-02) and EMBRC-France (ANR-10-INBS-02). Funding for the collection and processing of the Tara Oceans data set was provided by the NASA Ocean Biology and Biogeochemistry Program under grants NNX11AQ14G, NNX09AU43G, NNX13AE58G and NNX15AC08G (to the University of Maine), the Canada Excellence Research Chair in Remote Sensing of Canada’s New Arctic Frontier and the Canada Foundation for Innovation. The authors also thank agnès b. and E. Bourgois, the Prince Albert II de Monaco Foundation, the Veolia Foundation, Region Bretagne, Lorient Agglomeration, Serge Ferrari, Worldcourier and KAUST for support and commitment. The global sampling effort was made possible by countless scientists and crew who performed sampling aboard the Tara from 2009 to 2013, and the authors thank MERCATOR-CORIOLIS and ACRI-ST for providing daily satellite data during the expeditions. The authors are also grateful to the countries that graciously granted sampling permission. The authors thank N. Le Bescot and N. Henry for their help in designing the figures in this article. C.d.V. thanks the Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr). S. Sunagawa thanks the European Molecular Biology Laboratory and ETH Zürich’s high-performance computing facilities for computational support. C.B. acknowledges funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement 835067) as well as the Radcliffe Institute of Advanced Study at Harvard University for a scholar’s fellowship during the 2016–2017 academic year. M.B.S. thanks the Gordon and Betty Moore Foundation (award 3790) and the US National Science Foundation (awards OCE#1536989 and OCE#1829831) as well as the Ohio Supercomputer for computational support. S.G.A. thanks the Spanish Ministry of Economy and Competitiveness (CTM2017-87736-R). F.L. thanks the Institut Universitaire de France as well as the EMBRC platform PIQv for image analysis. S. Sunagawa is supported by ETH Zürich and the Helmut Horten Foundation and by funding from the Swiss National Foundation (205321_184955). The authors declare that all data reported herein are fully and freely available from the date of publication, with no restrictions, and that all of the analyses, publications and ownership of data are free from legal entanglement or restriction by the various nations in whose waters the Tara Oceans expeditions conducted sampling. This article is contribution number 100 of Tara Oceans.
Author information
Authors and Affiliations
Consortia
Contributions
S. Sunagawa and C.d.V. are the lead authors of the article and all other authors contributed to discussion of the content, writing and editing of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks David Hutchins, Maria Pachiadaki and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Artist profiles: https://oceans.taraexpeditions.org/en/m/art/artists/
Plankton chronicles: http://planktonchronicles.org
Tara Ocean Foundation: https://oceans.taraexpeditions.org/en
TaraOceans Sample Registry: https://doi.pangaea.de/10.1594/PANGAEA.875582
TaraOceans Sequencing: https://www.ebi.ac.uk/ena/data/view/PRJEB402
UniEuk: http://unieuk.org
Supplementary information
Glossary
- Epipelagic
-
Referring to uppermost layer of the ocean that receives sunlight, enabling the organisms inhabiting it to perform photosynthesis.
- Mesopelagic
-
Referring to the ocean layer that receives very little to no sunlight, lying beneath the epipelagic layer, ranging from about 200 to 1,000 m in depth.
- Heterotrophic
-
Capable of incorporating organic carbon into biomass.
- Remineralized
-
Derived from the breakdown of organic matter into its simplest inorganic form.
- Mixotrophy
-
Capacity to incorporate carbon into biomass from either inorganic or organic sources.
- Photoheterotrophy
-
Capacity to derive energy from light and carbon from organic matter.
- Haptophytes
-
Group of single-celled photosynthetic planktonic organisms.
- Metagenome-assembled genomes
-
(MAGs). Consensus genome sequences that are reconstructed using sequencing reads of DNA extracted from whole microbial communities.
- Eocene epoch
-
Second geological epoch of the Palaeogene period (66 million to 23 million years ago) that began 56 million years ago and ended 34 million years ago.
- Southern Ocean gateways
-
Pathways of the oceanic circulation that are influenced by the displacement of continents (for example, the Drake Passage, South African gateway and the Tasman gateway between Antarctica and South America, Africa and Australia, respectively).
- Bacterivory
-
Organisms that obtain carbon and energy primarily from the consumption of bacteria.
- Miocene epoch
-
First geological epoch of the Neogene period (2.6 million to 23 million years ago) that extends from about 23 million to 5 million years ago.
- Agulhas choke point
-
Oceanic system south of Africa where warm and salty Indian Ocean waters leak into the South Atlantic Ocean impacting the global oceanic circulation.
Rights and permissions
About this article
Cite this article
Sunagawa, S., Acinas, S.G., Bork, P. et al. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol 18, 428–445 (2020). https://doi.org/10.1038/s41579-020-0364-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-0364-5
This article is cited by
-
The road forward to incorporate seawater microbes in predictive reef monitoring
Environmental Microbiome (2024)
-
A genome and gene catalog of the aquatic microbiomes of the Tibetan Plateau
Nature Communications (2024)
-
Indexing and real-time user-friendly queries in terabyte-sized complex genomic datasets with kmindex and ORA
Nature Computational Science (2024)
-
Genome-scale community modelling reveals conserved metabolic cross-feedings in epipelagic bacterioplankton communities
Nature Communications (2024)
-
Datascape: exploring heterogeneous dataspace
Scientific Reports (2024)