Abstract
The end of the RNA polymerase II (Pol II) transcription cycle is strictly regulated to prevent interference between neighbouring genes and to safeguard transcriptome integrity1. The accumulation of Pol II downstream of the cleavage and polyadenylation signal can facilitate the recruitment of factors involved in mRNA 3′-end formation and termination2, but how this sequence is initiated remains unclear. In a chemical–genetic screen, human protein phosphatase 1 (PP1) isoforms were identified as substrates of positive transcription elongation factor b (P-TEFb), also known as the cyclin-dependent kinase 9 (Cdk9)–cyclin T1 (CycT1) complex3. Here we show that Cdk9 and PP1 govern phosphorylation of the conserved elongation factor Spt5 in the fission yeast Schizosaccharomyces pombe. Cdk9 phosphorylates both Spt5 and a negative regulatory site on the PP1 isoform Dis24. Sites targeted by Cdk9 in the Spt5 carboxy-terminal domain can be dephosphorylated by Dis2 in vitro, and dis2 mutations retard Spt5 dephosphorylation after inhibition of Cdk9 in vivo. Chromatin immunoprecipitation and sequencing analysis indicates that Spt5 is dephosphorylated as transcription complexes traverse the cleavage and polyadenylation signal, concomitant with the accumulation of Pol II phosphorylated at residue Ser2 of the carboxy-terminal domain consensus heptad repeat5. A conditionally lethal Dis2-inactivating mutation attenuates the drop in Spt5 phosphorylation on chromatin, promotes transcription beyond the normal termination zone (as detected by precision run-on transcription and sequencing6) and is genetically suppressed by the ablation of Cdk9 target sites in Spt5. These results suggest that the transition of Pol II from elongation to termination coincides with a Dis2-dependent reversal of Cdk9 signalling—a switch that is analogous to a Cdk1–PP1 circuit that controls mitotic progression4.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Proudfoot, N. J. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352, aad9926 (2016).
Davidson, L., Muniz, L. & West, S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 28, 342–356 (2014).
Sansó, M. et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 30, 117–131 (2016).
Grallert, A. et al. A PP1–PP2A phosphatase relay controls mitotic progression. Nature 517, 94–98 (2015).
Shetty, A. et al. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol. Cell 66, 77–88.e75 (2017).
Booth, G. T., Wang, I. X., Cheung, V. G. & Lis, J. T. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 26, 799–811 (2016).
Blethrow, J. D., Glavy, J. S., Morgan, D. O. & Shokat, K. M. Covalent capture of kinase-specific phosphopeptides reveals Cdk1–cyclin B substrates. Proc. Natl Acad. Sci. USA 105, 1442–1447 (2008).
Ohkura, H., Kinoshita, N., Miyatani, S., Toda, T. & Yanagida, M. The fission yeast dis2 + gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell 57, 997–1007 (1989).
Yamano, H., Ishii, K. & Yanagida, M. Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J. 13, 5310–5318 (1994).
Kinoshita, N., Ohkura, H. & Yanagida, M. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63, 405–415 (1990).
Viladevall, L. et al. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell 33, 738–751 (2009).
Pei, Y. & Shuman, S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J. Biol. Chem. 278, 43346–43356 (2003).
Sansó, M. et al. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet. 8, e1002822 (2012).
Booth, G. T., Parua, P. K., Sansó, M., Fisher, R. P. & Lis, J. T. Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nat. Commun. 9, 543 (2018).
Sajiki, K. et al. Genetic control of cellular quiescence in S. pombe. J. Cell Sci. 122, 1418–1429 (2009).
Cho, E. J., Kobor, M. S., Kim, M., Greenblatt, J. & Buratowski, S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15, 3319–3329 (2001).
Hausmann, S. & Shuman, S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 277, 21213–21220 (2002).
Schwer, B., Ghosh, A., Sanchez, A. M., Lima, C. D. & Shuman, S. Genetic and structural analysis of the essential fission yeast RNA polymerase II CTD phosphatase Fcp1. RNA 21, 1135–1146 (2015).
St. Amour, C. V. et al. Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) Rpb1 carboxyl-terminal domain substrate recognition. Mol. Cell. Biol. 32, 2372–2383 (2012).
Pei, Y. et al. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol. Cell. Biol. 26, 777–788 (2006).
Vanoosthuyse, V. et al. CPF-associated phosphatase activity opposes condensin-mediated chromosome condensation. PLoS Genet. 10, e1004415 (2014).
Nedea, E. et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29, 577–587 (2008).
Schreieck, A. et al. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat. Struct. Mol. Biol. 21, 175–179 (2014).
Baejen, C. et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol Cell 66, 38–49.e36 (2017).
Bernecky, C., Herzog, F., Baumeister, W., Plitzko, J. M. & Cramer, P. Structure of transcribing mammalian RNA polymerase II. Nature 529, 551–554 (2016).
Schneider, S., Pei, Y., Shuman, S. & Schwer, B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell. Biol. 30, 2353–2364 (2010).
Fong, N. et al. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell 60, 256–267 (2015).
Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D. L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 15, 71–78 (2008).
Fong, N., Saldi, T., Sheridan, R. M., Cortazar, M. A. & Bentley, D. L. RNA Pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol. Cell 66, 546–557.e3 (2017).
Yamada, T. et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 (2006).
Hayashi, A. et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells 14, 217–225 (2009).
Saiz, J. E. & Fisher, R. P. A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr. Biol. 12, 1100–1105 (2002).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991).
Stone, E. M., Yamano, H., Kinoshita, N. & Yanagida, M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 3, 13–26 (1993).
Parua, P. K., Mondal, A. & Parrack, P. HflD, an Escherichia coli protein involved in the λ lysis–lysogeny switch, impairs transcription activation by λCII. Arch. Biochem. Biophys. 493, 175–183 (2010).
Sansó, M. et al. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 39, 6369–6379 (2011).
Tanny, J. C., Erdjument-Bromage, H., Tempst, P. & Allis, C. D. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21, 835–847 (2007).
Kao, C. F. & Osley, M. A. In vivo assays to study histone ubiquitylation. Methods 31, 59–66 (2003).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP–seq enrichment using MACS. Nat. Protocols 7, 1728–1740 (2012).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Mahat, D. B. et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO–seq). Nat. Protocols 11, 1455–1476 (2016).
Acknowledgements
R.P.F. is grateful for the mentorship provided by Günter Blobel (1936–2018). We thank I. M. Hagan, B. Schwer, S. Shuman, V. Vanoosthuyse, M. Yanagida, M. J. O’Connell and the National BioResource Project/Yeast Genetic Resource Center for providing yeast strains and/or antibodies; K. M. Shokat for providing 3-MB-PP1; C. Zhang for guidance in analogue-sensitive allele optimization; D. Hasson for advice on ChIP–seq data analysis and N. Steinbach and R. Parsons for assistance in phosphatase-activity measurements. J.C.T. was supported by Canadian Institutes of Health Research grant MOP-130362 and by a fellowship from Fond de recherche Quebec Santé (3315). This work was supported by National Institutes of Health grants GM25232 to G.T.B. and J.T.L. and GM104291 to R.P.F. Next-generation sequencing was supported in part by grant P30 CA196521 to the Tisch Cancer Institute.
Author information
Authors and Affiliations
Contributions
P.K.P. and M.S. identified PP1 isoforms as Cdk9 substrates. P.K.P. conducted enzymologic studies, measured Spt5 dephosphorylation rates in wild-type and PP1 mutant backgrounds, performed ChIP–qPCR analysis and characterized genetic interactions between dis2 and spt5 mutant alleles. P.K.P., M.S., B.B. and J.C.T. performed ChIP–seq analysis to map distribution of pSpt5. G.T.B. performed PRO–seq analysis and developed metrics to quantify termination defects in dis2 mutants. P.K.P., G.T.B., J.T.L. and R.P.F. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 A Cdk9–Dis2–Spt5 regulatory circuit.
a, Cdk9-dependence of Dis2-T316 phosphorylation in vivo. Cells of wild-type (wt) or cdk9as strains, with or without GFP-tagged Dis2 expressed from the chromosomal dis2+ locus, were treated for 10 min with 20 μM 3-MB-PP1 or mock-treated, as indicated. Chromatin extracts were immunoprecipitated with anti-GFP antibodies and probed with antibodies specific for Dis2 phosphorylated at Thr316 (Dis2-pT316) or GFP (n = 2 independent repeats). b, Spt5 dephosphorylation by purified PP1 in vitro. Purified GST–Dis2 and GST–Sds21 were incubated with a control phosphopeptide derived from histone H3 (H3pS10), an Spt5 CTD consensus phosphopeptide (Spt5-pT1) or a non-phosphorylated peptide of the same sequence (Spt5-NP). c, Spt5 dephosphorylation by PP1 isolated from fission yeast. A polyclonal anti-Dis2 antibody immunoprecipitates an active pSpt5 phosphatase from extracts of dis2+ but not dis2Δ or dis2-11 mutant cells. Note, the antibody cross-reacts with Sds21 in immunoblots but does not efficiently immunoprecipitate Sds21. d, Loss of Dis2 activity upon Cdk9-dependent phosphorylation. As in Fig. 1d, results show activity of Dis2 (isolated from yeast and phosphorylated by Cdk9 or mock-treated) towards H3pS10. e, Top, anti-Myc immunoprecipitates from Myc–Dis2-expressing cells were tested for phosphatase activity towards Spt5-pT1 and H3pS10 peptides. Bottom, immunoblot to verify expression and immunoprecipitation of Myc–Dis2. b–e, Data are mean + s.d. from three biological replicates.
Extended Data Fig. 2 Distinct kinase–phosphatase circuits regulate phosphorylation of Spt5 Thr1 and Rpb1 Ser2 in vivo.
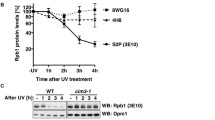
a, Rapid dephosphorylation of Spt5 after Cdk9 inhibition and stabilization of pSpt5 by Dis2 inactivation occur in the spt5+ strain and do not depend on the C-terminal Myc-epitope tag. b, Spt5 dephosphorylation kinetics in single, double and triple cdkas mutants treated with 3-MB-PP1 show that Cdk9 is the sole kinase needed to phosphorylate this site in vivo. c, Fcp1 inactivation stabilizes Rpb1 Ser2 phosphorylation after Lsk1 inhibition. Fission-yeast strains, lsk1as or lsk1as fcp1-452, were grown at 30 °C and shifted to 37 °C (or not shifted), treated for the indicated time with 20 μM 3-MB-PP1, and analysed by immunoblotting for Pol II Ser2 phosphorylation. Note, CTD dephosphorylation leads to increased reactivity with 8WG16 antibody used to detect total Pol II. d, Dis2 activity is dispensable for Pol II CTD Ser2 dephosphorylation. As in Fig. 2a, except that the experiment was performed in lsk1as cells, 20 μM 3-MB-PP1 was added and extracts were probed for Pol II Ser2 phosphorylation (or tubulin as a loading control). e, Fcp1 activity is dispensable for Spt5 Thr1 dephosphorylation. As in c, except that strains carried a cdk9as allele and were tested for both pSpt5 (unaffected by Fcp1 inactivation) and pSer2 (unaffected by Cdk9 inhibition). f, CPF-associated Pol II pSer5 phosphatase Ssu72 is dispensable for Spt5 Thr1 dephosphorylation. Fission-yeast strains (cdk9as, cdk9as ssu72C13S and ssu72C13S) were grown at 30 °C, treated for the indicated time with 10 μM 3-MB-PP1 and analysed by immunoblotting for pSpt5 and total Spt5. a–f, n = 2 independent repeats.
Extended Data Fig. 3 Chromatin-associated Spt5 is dephosphorylated rapidly upon Cdk9 inhibition and stabilized in dis2-11 cells.
a, Rapid pSpt5 turnover on chromatin. ChIP–qPCR analysis of crosslinking of pSpt5 versus total Spt5 at the eng1+ gene after 3-MB-PP1 treatment for various lengths of time. Left, absolute ChIP signals for anti-Myc; middle, absolute signals for anti-pSpt5; right, ratio of pSpt5 to total Spt5, expressed as a percentage of the ratio in the absence of the inhibitor. b, Loss of Dis2 function stabilizes pSpt5 on chromatin. Left and middle, either cdk9as spt5-13Myc dis2+ or cdk9as spt5-13Myc dis2-11 cells were shifted to 18 °C and treated with 10 μM 3-MB-PP1 or mock-treated with DMSO for 2 min and subjected to ChIP–qPCR analysis at the eng1+ locus for Spt5–Myc (left) or pSpt5 (middle). Right, the pSpt5:Spt5 signal ratios of 3-MB-PP1-treated samples versus DMSO-treated samples. The pSpt5:Spt5 signal ratios between treatments (DMSO and 3-MB-PP1) were plotted for each condition (dis2+ or dis2-11). Note, higher residual levels of pSpt5 in cdk9as dis2+ cells, compared to those analysed in a, may reflect less efficient dephosphorylation at 18 °C, relative to 30 °C. c, As in b, except measuring at the aro1+ gene. d, As in b, but measuring at the hxk2+ gene (raw data for pSpt5 and total Spt5 from which ratios in Fig. 2c were calculated). a–d, Data are mean + s.d. from three biological replicates. b–d, P values (Student’s t-test) are indicated between wild-type (dis2+) and mutant (dis2-11) cells.
Extended Data Fig. 4 A specific link between Cdk9 activity and Dis2 recruitment to chromatin.
a, Increased recruitment of Dis2 to chromatin is a specific consequence of Cdk9 inhibition. ChIP–qPCR analysis of Myc–Dis2 (left), Pol II (middle) and Myc–Dis2:Pol II signal ratios (right) at the eng1+ locus in cdk9as cells, expressing Myc–Dis2 from a plasmid, treated with 20 μM 3-MB-PP1 or DMSO for 10 min at 30 °C. b, c, Mcs6 and Lsk1 activities do not influence Dis2 recruitment to chromatin. As in a, except cells containing mcs6as5 (b) or lsk1as (c) alleles were treated with 20 μM 3-MB-PP1 for 10 min at 30 °C. In a–c, data are mean + s.d. from three biological replicates. d, Constitutive cdk9 loss-of-function mutations increase GFP–Dis2 recruitment to chromatin. Dis2 occupancy at the eng1+ locus analysed in cdk9+ cells, a cdk9ΔC mutant and a cdk9T212A mutant. Data are mean + s.d. from technical duplicates of one experiment.
Extended Data Fig. 5 Regulation of pSpt5 by Cdk9 and Dis2 occurs independently of CPF recruitment and upstream of CPF function.
a, Core CPF recruitment to chromatin is unaffected by Cdk9 inhibition. Cells with different GFP-tagged CPF subunits expressed from their respective chromosomal loci (cdk9as pfs2–GFP–HA, cdk9as pla1–GFP–HA or cdk9as cft1–GFP) were grown at 30 °C and treated with 10 μM 3-MB-PP1 or DMSO for 10 min. ChIP–qPCR analysis of GFP:Pol II signal ratios was performed at the hxk2+ gene. Data are mean + s.d. from three biological replicates. b, Wild-type (dis2+) and mutant (dis2-11) cells were grown at 30 °C and shifted to 18 °C (or not shifted) for 10 min. ChIP–qPCR analysis of Pol II-pTyr1:Pol II (top), pSpt5:Spt5 (middle) and Pol II-pSer2:Pol II (bottom) signal ratios was performed at the psu1+ gene. c, Loss of Dis2 activity does not affect chromatin recruitment of a core CPF subunit, Pfs2. Cells of dis2+ and dis2-11 strains with Pfs2–GFP–HA expressed from the chromosomal pfs2+ locus were grown at 30 °C and shifted to 18 °C (or not shifted) for 10 min before formaldehyde crosslinking and chromatin isolation. ChIP–qPCR analysis of GFP:Pol II signal ratios was performed at the psu1+ gene at 30 °C (top) and 18 °C (bottom). d, Spt5 phosphorylation is not affected by thermal inactivation of an essential CPF subunit. Cells of pfs2+ and pfs2-11 (temperature-sensitive) strains were grown at 30 °C, shifted to 37 °C (or not shifted) and incubated for various times as indicated. ChIP–qPCR analysis of pSpt5, total Spt5, pSpt5:Spt5 signal ratios, Pol II and Pol II signal ratios (pfs2-11 over pfs2+), was performed at the hxk2+ locus. b–d, Data are mean + s.d. from two biological replicates.
Extended Data Fig. 6 PP1 allele- and isoform-specific stabilization of Spt5 phosphorylation on chromatin.
a, ChIP–qPCR analysis at the rps17a+ gene. Comparison of pSpt5:Spt5 ratio in the indicated strains upstream and downstream of the CPS at 30 °C (left) and comparison of the ratio between dis2+ and dis2-11 cells at 18 °C (right). b, ChIP–qPCR analysis at the hxk2+ gene. Comparison of the pSpt5:Spt5 ratio in the indicated strains upstream and downstream of the CPS at 30 °C. a, b, Data are mean + s.d. from three biological replicates; P values (Student’s t-test) between wild-type (dis2+) and mutants (dis2Δ, dis2-11, dis2T316A or dis2T316D) are indicated. c, Dephosphorylation of Spt5 after Cdk9 inhibition is retarded in a dis2Δ strain, relative to a dis2+ strain. d, Spt5-dephosphorylation kinetics after Cdk9 inhibition are unaffected by sds21 deletion in a dis2+ strain. c, d, n = 2 independent repeats. e, Cdk9 does not restrict chromatin recruitment of Sds21. Anti-GFP ChIP–qPCR analysis at the eng1+ locus in a cdk9as GFP–sds21 strain treated for 10 min with 10 μM 3-MB-PP1 reveals unchanged (or slightly decreased) Sds21 occupancy when Cdk9 is inhibited. Data are mean + s.d. from technical duplicates of one experiment.
Extended Data Fig. 7 Spt5 and pSpt5 ChIP–seq analysis.
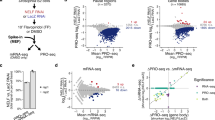
a, Correlation between ChIP–seq samples. Paired-end sequencing reads were mapped to the fission-yeast genome using Bowtie2 (Galaxy v.2.2.6.2). Mapped reads of each biological replicate were used to calculate correlation between pairs of replicates. Values in boxes represent Pearson’s correlation coefficients between corresponding samples (n = 2 biological replicates). b, Metagene (top) and heat map (bottom) analyses show genome-wide (n = 3,054 genes) comparison of pSpt5:Spt5 ratios (log2) between dis2+ and dis2-11 cells (raw data from which fold change in Fig. 3c was calculated). c, Metagene plots (top) and heat maps (bottom) show Spt5–Myc and pSpt5 distribution in dis2+ and dis2-11 cells, as indicated, across Pol II-transcribed genes (n = 175), filtered to include only genes separated from nearest neighbours by more than 500 bp at both ends, on both strands. d, Metagene plot (top) and heat map (bottom) represent fold-change of pSpt5:Spt5 ratio in dis2-11 over dis2+ around CPS of the genes analysed in c. In b, c, regions between +300 bp relative to the TSS and −300 bp relative to the CPS were scaled to enable comparison of genes of different lengths.
Extended Data Fig. 8 Relative distributions of Pol II and Spt5.
a, Pol II distribution on chromatin. Metagene plot (top) and heat map (bottom) of Pol II ChIP–seq distributions (data from previously published work5) across Pol II-transcribed genes (n = 3,054) in wild-type cells. Genes were sorted on the basis of Pol II occupancy and k-means clustering was performed to partition genes into three clusters with nearest median occupancy. b, Pol II pSer2 distribution on chromatin. Metagene plot (top) and heat map (bottom) of Pol II pSer2 (data from previously published work5) across Pol II-transcribed genes (n = 3,054) in wild-type cells. c, Distribution of pSpt5 and pSer2 on chromatin. Metagene analyses (top) and heat maps (bottom) show genome-wide comparison between log2 ratios of pSpt5:Spt5 and pSer2:Pol II for Pol II-transcribed genes (n = 3054) in wild-type cells (separate plots of data superimposed in Fig. 3d). In a–c, regions between +300 bp relative to the TSS and −300 bp relative to the CPS were scaled to enable comparison of genes of different lengths. d, Spt5 and pSpt5 distribution around CPS of Pol II-transcribed genes. Metagene plots (top) and heat maps (bottom) represent the distribution of Spt5–Myc and pSpt5 around the CPS (−500 to +500 bp) of highly active genes (n = 137; cluster 1 of Fig. 3a in dis2+ and dis2-11 cells. e, Statistical analysis of Spt5–Myc and pSpt5 occupancy around CPS. Box plots represent occupancy of Spt5 and pSpt5 in the region 500 bp downstream of the CPS (y; PostCPS) versus the region 500 bp upstream of the CPS (x; PreCPS), for genes in cluster 1 of Fig. 3a (n = 137), in dis2+ and dis2-11 cells (Spt5, dis2+: high = 3.506, low = 5.5312 × 10−2, median = 0.9199, 95% confidence interval; pSpt5, dis2+: high = 2.085, low = 2.8570 × 10−2, median = 0.4655, 95% confidence interval; Spt5, dis2-11: high = 1.617, low = 5.7597 × 10−2, median = 0.6042, 95% confidence interval; pSpt5, dis2-11: high = 1.155, low = 6.2344 × 10−2, median = 0.4530, 95% confidence interval). f, Box plots represent statistical significance of increases in pSpt5:Spt5 ratios in PostCPS versus PreCPS regions in dis2-11 versus dis2+ cells (n = 137) (dis2+: high = 1.533, low = 4.5765 × 10−2, median = 0.5356, 95% confidence interval; dis2-11: high = 1.601, low = 0.4651, median = 0.7482, 95% confidence interval). e, f, P values were calculated using two-sided Student’s t-test. g, An Spt5 that cannot be phosphorylated suppresses conditional lethality of dis2-11. Growth kinetics in liquid culture of indicated strains after a shift to 18 °C. Data are mean ± s.d. from two biological replicates.
Extended Data Fig. 9 The dis2-11 mutation affects global transcription properties independent of temperature.
a, PRO–seq experiments are reproducible. Scatter plots comparing PRO–seq libraries from two biological replicates for each experiment. Values represent log10(normalized reads) within the gene body (TSS +200 bp to the CPS) of all filtered genes (n = 3383). Colours indicate the numbers of genes represented by each point. Normalization on the basis of spike-in should centre scatter about the diagonal line x = y (magenta, dotted). Correlation values represent Spearman’s rank correlation. b, Comparison of composite PRO–seq profiles of dis2-11 mutant alone (top panels) or cdk9as dis2-11 (bottom panels) at 18 °C and 30 °C. Profiles are centred either on the TSS (left) or the CPS (right). Shaded areas on composite profiles represent the 12.5 and 87.5% quantiles at each position. c, Composite PRO–seq profiles comparing dis2+ strain (cdk9as) with dis2-11 strain, both at 18 °C. Genes were scaled to a common length by fixing the middle gene body region (TSS + 300 bp to CPS − 300 bp) to 60 windows. b, c, Solid lines represent an averaged-data plot and shaded regions represent s.d. of the median. d, Heat maps of spike-in normalized PRO–seq signal (log10) within 10-bp windows relative to the CPS (−250 to +1,000) for cdk9as (left) and cdk9as dis2-11 (right) strains at 18 °C. Genes were ranked by decreasing TEI in cdk9as dis2-11 at 18 °C, a measure of the termination-window size. Each panel represents data from filtered genes that are at least 1 kb from neighbouring genes on the same strand (n = 939). In a–d, data are from two biological replicates.
Extended Data Fig. 10 Multiple dis2 mutations cause termination defects.
a, Browser image displaying normalized PRO–seq signal at the SPAC1002.13c gene locus. Track values reflect the maximum displayed signal (some peaks exceed these values). b, Box plots displaying the distribution of TEI values in each strain for all filtered genes separated from same-strand neighbours by at least 1 kb (n = 939). Significant differences (P values from Welch’s two sample t-test) in mean TEI for each strain compared with dis2+ are indicated (dis2+: high = 1.2304, low = −2.4005, median = −0.6532; dis2-11: high = 1.0066, low = −2.3483, median = −0.3802; dis2Δ: high = 1.5563, low = −1.8325, median = −0.06695; dis2T316A: high = 1.4314, low = −2.1004, median = −0.1176; dis2T316D: high = 1.3424, low = −2.0934, median = −0.2310; each box shows 25th–75th percentiles). c, Composite PRO–seq profiles of each dis2 mutant strain (red) compared with dis2+ (blue). Profiles reflect the region from −250 bp to +1,000 bp around the CPS. Shaded areas on composite profiles represent the 12.5 and 87.5% quantiles at each position. Each panel represents data from filtered genes that are at least 1 kb from neighbouring genes on the same strand (n = 939). Solid lines represent an averaged-data plot of the median. d, Heat maps displaying log2(mutant/wild-type) PRO–seq signal within 10-bp windows from −250 bp to +1,000 bp around the CPS for all genes used in c sorted by decreasing TEI values in dis2+ (top to bottom). In a–d, data are from two biological replicates.
Supplementary information
Supplementary Figures
This file contains the uncropped images of the Phosphorimager scans, stained gels and immunoblots presented in Figs. 1b-d, 2a, b and Extended Data Figs. 1a, c, e, 2a-f and 6c, d.
Supplementary Table 1
This file contains a list of fission yeast strains used in this study.
Supplementary Table 2
This file contains a list of DNA oligonucleotide primers used in this study.
Rights and permissions
About this article
Cite this article
Parua, P.K., Booth, G.T., Sansó, M. et al. A Cdk9–PP1 switch regulates the elongation–termination transition of RNA polymerase II. Nature 558, 460–464 (2018). https://doi.org/10.1038/s41586-018-0214-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0214-z
This article is cited by
-
Significance of hepatitis B virus capsid dephosphorylation via polymerase
Journal of Biomedical Science (2024)
-
Structural basis of exoribonuclease-mediated mRNA transcription termination
Nature (2024)
-
The NELF pausing checkpoint mediates the functional divergence of Cdk9
Nature Communications (2023)
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex
Nature Reviews Molecular Cell Biology (2023)
-
MYC promotes global transcription in part by controlling P-TEFb complex formation via DNA-binding independent inhibition of CDK9 SUMOylation
Science China Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.