Abstract
In mammals, 2′-O-methylation of RNA is a molecular signature by which the cellular innate immune system distinguishes endogenous from exogenous messenger RNA1,2,3. However, the molecular functions of RNA 2′-O-methylation are not well understood. Here we have purified TAR RNA-binding protein (TRBP) and its interacting partners and identified a DICER-independent TRBP complex containing FTSJ3, a putative 2′-O-methyltransferase (2′O-MTase). In vitro and ex vivo experiments show that FTSJ3 is a 2′O-MTase that is recruited to HIV RNA through TRBP. Using RiboMethSeq analysis4, we identified predominantly FTSJ3-dependent 2′-O-methylations at specific residues on the viral genome. HIV-1 viruses produced in FTSJ3 knockdown cells show reduced 2′-O-methylation and trigger expression of type 1 interferons (IFNs) in human dendritic cells through the RNA sensor MDA5. This induction of IFN-α and IFN-β leads to a reduction in HIV expression. We have identified an unexpected mechanism used by HIV-1 to evade innate immune recognition: the recruitment of the TRBP–FTSJ3 complex to viral RNA and its 2′-O-methylation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RiboMethSeq data that support the findings of this study have been deposited in the European Nucleotide Archive (ENA) with the accession code PRJEB29444.
References
Züst, R. et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 (2011).
Daffis, S. et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456 (2010).
Schuberth-Wagner, C. et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2'O-methylated self RNA. Immunity 43, 41–51 (2015).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 44, e135 (2016).
Motorin, Y. & Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631 (2011).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Shuman, S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3, 619–625 (2002).
Werner, M. et al. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 39, 4756–4768 (2011).
David, R. Immune evasion: Gm18, a bacterial ‘invisibility cloak’. Nat. Rev. Microbiol. 10, 238–239 (2012).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Decroly, E., Ferron, F., Lescar, J. & Canard, B.Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10, 51–65 (2011).
Dong, H. et al. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 8, e1002642 (2012).
Martin, B., Reynard, O., Volchkov, V. & Decroly, E. Filovirus proteins for antiviral drug discovery: structure/function of proteins involved in assembly and budding. Antiviral Res. 150, 183–192 (2018).
Coutard, B. et al. Zika virus methyltransferase: structure and functions for drug design perspectives. J. Virol. 91, e02202-16 (2017).
Gehrig, S. et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 (2012).
Jöckel, S. et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 (2012).
Gatignol, A., Buckler-White, A., Berkhout, B. & Jeang, K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251, 1597–1600 (1991).
Benkirane, M. et al. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 16, 611–624 (1997).
Hodel, A. E., Gershon, P. D., Shi, X. & Quiocho, F. A. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 85, 247–256 (1996).
Lapeyre, B. & Purushothaman, S. K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell 16, 663–669 (2004).
Chendrimada, T. P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Ciuffi, A. Viral cell biology: HIV RNA gets methylated. Nat. Microbiol. 1, 16037 (2016).
Blanco-Melo, D., Venkatesh, S. & Bieniasz, P. D. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 37, 399–411 (2012).
Laguette, N. & Benkirane, M. How SAMHD1 changes our view of viral restriction. Trends Immunol. 33, 26–33 (2012).
Nakatani, Y. & Ogryzko, V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370, 430–444 (2003).
Bennasser, Y. et al. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat. Struct. Mol. Biol. 18, 323–327 (2011).
Bélanger, F., Stepinski, J., Darzynkiewicz, E. & Pelletier, J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 285, 33037–33044 (2010).
Martin, B. et al. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res. 46, 7902–7912 (2018).
Grosjean, H., Droogmans, L., Roovers, M. & Keith, G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 425, 55–101 (2007).
Bauer, D. E., Canver, M. C. & Orkin, S. H. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. 95, e52118 (2015).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Rasaiyaah, J. et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 (2013).
Watts, J. M. et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 (2009).
Brussel, A. & Sonigo, P. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J. Virol. 78, 11263–11271 (2004).
Acknowledgements
We thank M. Benkirane and members of the Molecular Virology Laboratory for support and reading of the manuscript. This work was supported by grants from the European FP7 contract 201412, MSDAvenir, and FRM ‘équipe labellisée’ to M. Benkirane, head of the Molecular Virology Laboratory. This work was also supported by an ANR HTRNAMod (ANR-13-ISV8-0001) grant to Y.M. M.R. was supported by ‘Région Languedoc Roussillon’.
Reviewer information
Nature thanks P. Y. Shi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Y.B. conceived the study. Y.B. and M.R. designed experiments and interpreted data. V.M. and Y.M. performed RiboMethSeq experiments, and analysed and interpreted data. E.D. performed internal methylation assay experiments. Y.B. wrote the paper. All the authors discussed the data, read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
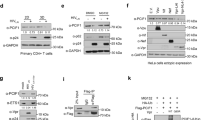
Extended Data Fig. 1 FTSJ3 is a human 2′-O-methyltransferase.
a, Reciprocal immunoprecipitation was performed on HeLa S3 cells expressing Flag/HA epitope-tagged FTSJ3 protein (FTSJ3–FH). Eluates were analysed using anti-HA and anti-TRBP antibodies. The experiment was repeated four times with similar results. b, FTSJ3 methylation activity was assessed in vitro using TAR RNA as a substrate. Radiolabelled 32P-TAR was incubated for 1 h at 30°C with equal amounts of Flag/HA-immunoprecipitated FSTJ3–FH or mock immunoprecipitate blotted in Fig. 1d. RNAs were then treated with nuclease P1 for 3 h, and released caps were analysed by TLC chromatography migration using 0.3 M ammonium sulfate buffer. The plate was dried and exposed by autoradiography. The experiment was repeated twice with similar results. c, Products from in vitro mock and FTSJ3–FH TAR methylation were digested with TAP. As a control, 32P-m7G-TAR was also digested with TAP, to release the m7GMP residue. Products were analysed on TLC plates developed with 0.45 M ammonium sulfate buffer. The experiment was repeated twice with similar results. d, Purified recombinant FTSJ3 was incubated without exogenous RNA (–) or with the homopolymeric synthetic 27-mer RNAs A27, U27, G27 and C27, and associated MTase activity was determined by [3H]-methyl incorporation assay. The background obtained in the absence of exogenous RNA was subtracted. n = 3 independent samples, results shown as the mean ± s.d. representative of three experiments. Indicated P values obtained using two-tailed Student’s t-test.
Extended Data Fig. 2 RiboMethSeq analysis of 2′-O-methylated sites detected in HIV RNA isolated from viral particles.
RiboMethSeq data are presented for each of the seventeen 2′-O-methylated sites. Data are superposed with relative local cleavage profiles for in vitro T7 transcripts of HIV-1 RNA, WT-HIV-1 RNA, siFTSJ3-HIV-1 RNA and CRISPR–Cas9-FTSJ3-HIV-1 RNA extracted from virions. Individual points for biological replicates are shown; n = 2 or n = 3. Position numbers are indicated on the bottom. In RiboMethSeq, the protection caused by 2′-O-methylation appears at the 3′-neighbouring position in RNA. For simplicity, a shift of –1 was applied to numbering here, so that the increased protection signal falls exactly to the 2′-O-methylated nucleotide.
Extended Data Fig. 3 RiboMethSeq analysis of 2′-O-methylated sites detected in HIV RNA isolated from viral particles: scores.
Variations and absolute values of RiboMethSeq scores (ScoreC, ScoreMean, ScoreA and ScoreB) for each of the seventeen methylated positions. ScoreC, ScoreMean and ScoreA can vary from 0 to 1 (negative values are possible), whereas Score B may be >1 for highly methylated positions in RNA. Individual points for biological replicates are shown; n = 2 or n = 3.
Extended Data Fig. 4 Validation of 783A, 1423U, 8063A, 8403A and 8407A 2′-O-methylation using primer extension assay.
HIV-1 RNA isolated from viral particles produced in mock cells (WT) or cells knocked-out for FTSJ3 using CRISPR–Cas9 (CRISPR–Cas9-FTSJ3) were subjected to primer extension assay using specific radiolabelled primers. Extension assay was realized at high (lanes 1, 3) and low concentrations (lanes 2, 4) of dNTP as indicated. The sequencing experiment was run side-by-side using WT-HIV RNA as a template (lanes 4–8). Primer extension products were analysed on an acrylamide–urea sequencing gel and visualized by autoradiography. Arrows, 2′-O-methylated position and stop. Results are representative of two independent experiments.
Extended Data Fig. 5 Characterization of HIV-1 RNA transfected in U937 cells.
a, HIV viruses were produced in 293T cells transfected with FTSJ3 siRNA or a control non-specific siRNA (scramble). Inhibition of FTSJ3 was assessed by western blot. The experiment was repeated more than 10 times with similar results or a more efficient knockdown. b, Purified recombinant wild-type or catalytic KDKE mutant FTSJ3–His was incubated with in vitro-transcribed HIV-1 RNA in the presence of radiolabelled [3H]-SAM, and MTase activity was determined by [3H]-methyl incorporation. n = 3 independent samples; results shown as the mean ± s.d. representative of two independent experiments. Indicated P values obtained by two-tailed Student’s t-test. c, Purified recombinant WT-FTSJ3 was incubated without exogenous RNA or with the 8362–8461 HIV RNA fragment and MTase activity was determined by [3H]-methyl incorporation assay (left) or 2D TLC separation (right). [3H]-methyl incorporation assay results were normalized by subtracting the background obtained in the absence of exogenous RNA. 2D TLC separation of 5′-[32P]-NMPs resulted from nuclease P1 hydrolysis of HIV RNA transcript 8362–8461 radiolabelled with [α-32P]ATP and incubated with WT-FTSJ3 or mutant KDKE-FTSJ3 recombinant proteins. Migration was performed in the N1/N2 or N1/R2 combination of solvents (see Methods). Positions of unmodified AMP (pA), 2′-O-methyl AMP (pAm) and inorganic phosphate (Pi) are indicated. The experiment was repeated twice with similar results.
Extended Data Fig. 6 HIV viral particles produced in TRBP knockdown cells induce type-1 interferon expression whereas viruses produced in siMRM2 knockdown and siFTSJ3-empty virus-like particles do not.
a, b, 293T cells were transfected with siRNA targeting TRBP (siTRBP) (a), FTSJ3 (siFTSJ3) or MRM2 (siMRM2) (b) or a control non-specific siRNA (scramble). After 16 h, cells were further transfected with pNL4-3 HIV molecular clone. Viruses were harvested 48 h later and quantified for RT, p24 and packaged viral RNA. Top, protein inhibition was assessed by western blot. U937 cells were infected with same amounts of viruses produced in cells treated with scramble siRNA (WT-HIV) or siTRBP, siFTSJ3 or siMRM2 as indicated. Bottom, 16 h later, expression of IFN-α and IFN-β was quantified by qRT–PCR. n = 3 biological samples, representative of two experiments. c, Empty virus-like particles were produced in scramble-293T cells or siFTSJ3-293T cells and used to transduce U937 cells. Expression of IFN-α and IFN-β was quantified by qRT–PCR 16 h later. n = 3 biological samples. Experiment repeated three times with similar results. d, RT-mutant HIV-BaL virus produced in scramble- or siFTSJ3-treated cells was used to infect U937 cells. Type-1 interferons were quantified by qRT–PCR 16 h after infection. Knockdown of FTSJ3 expression in HIV-producing cells was assessed by western blot (left). n = 3 biological samples representative of two independent experiments. Results shown as the mean ± s.d. Indicated P values obtained by two-tailed Student’s t-test.
Extended Data Fig. 7 Characterization of shMDA5 and shRIG-I knockdown U937 cells.
a, Inhibition of RIG-I using shRNA in U937 cells assessed by western blot. Experiment repeated three times with similar results. b, U937-shMDA5 and U937-shRIG-I cells were characterized for their ability to induce IFN-β after poly(I:C) treatment or Sendai virus infection. As indicated, n = 2 or n = 3 biological replicates representative of two experiments. Results shown as the mean ± s.d. Indicated P values obtained by two-tailed Student’s t-test. c, Type-1 interferon expression in U937 or U937-shRIG-I cells infected by siFTSJ3-HIV viral particles. IFN-α and IFN-β were quantified by qRT–PCR 16 h after infection. n = 2 independent samples, representative of three experiments. Results shown as the mean ± s.d. Indicated P values obtained by two-tailed Student’s t-test.
Extended Data Figure 8 siFTSJ3-HIV induces phosphorylation of IRF-3, IRF-7 and STAT1 transcriptional regulators and expression of type-1 interferons in MDMs.
a, Monocytes were purified from human peripheral blood mononuclear cells from healthy donors using CD14 selection magnetic beads. Monocytes were differentiated using GM-CSF for 5 days into macrophages and checked by FACS analysis. The same amounts of WT-HIV or siFTSJ3-HIV were used to infect primary cells. After 4 h, phosphorylation of IRF3, IRF7 and STAT1 was assessed by western blot. ERK-1/2 level was assessed as a loading control. Experiment repeated twice with similar results. b, Cells were harvested and RNAs purified 16 h post-infection. IFN-α and IFN-β were quantified using qRT–PCR and normalized to actin. Results are expressed as fold increase compared to uninfected cells. n = 3 biological replicates representative of two experiments realized on different donors, results shown as the mean ± s.d. Indicated P values obtained by two-tailed Student’s t-test.
Supplementary information
Supplementary Figures
This file contains uncropped scans with size marker indications.
Source data
Rights and permissions
About this article
Cite this article
Ringeard, M., Marchand, V., Decroly, E. et al. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504 (2019). https://doi.org/10.1038/s41586-018-0841-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0841-4
This article is cited by
-
Single-molecule epitranscriptomic analysis of full-length HIV-1 RNAs reveals functional roles of site-specific m6As
Nature Microbiology (2024)
-
Regulation and functions of non-m6A mRNA modifications
Nature Reviews Molecular Cell Biology (2023)
-
RNA modifications in cancer
British Journal of Cancer (2023)
-
Single-base resolution mapping of 2′-O-methylation sites by an exoribonuclease-enriched chemical method
Science China Life Sciences (2023)
-
Advances on genetic and genomic studies of ALV resistance
Journal of Animal Science and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.