Abstract
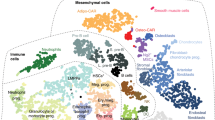
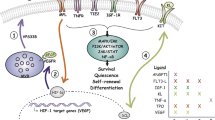
The bone marrow microenvironment has a key role in regulating haematopoiesis, but its molecular complexity and response to stress are incompletely understood. Here we map the transcriptional landscape of mouse bone marrow vascular, perivascular and osteoblast cell populations at single-cell resolution, both at homeostasis and under conditions of stress-induced haematopoiesis. This analysis revealed previously unappreciated levels of cellular heterogeneity within the bone marrow niche and resolved cellular sources of pro-haematopoietic growth factors, chemokines and membrane-bound ligands. Our studies demonstrate a considerable transcriptional remodelling of niche elements under stress conditions, including an adipocytic skewing of perivascular cells. Among the stress-induced changes, we observed that vascular Notch delta-like ligands (encoded by Dll1 and Dll4) were downregulated. In the absence of vascular Dll4, haematopoietic stem cells prematurely induced a myeloid transcriptional program. These findings refine our understanding of the cellular architecture of the bone marrow niche, reveal a dynamic and heterogeneous molecular landscape that is highly sensitive to stress and illustrate the utility of single-cell transcriptomic data in evaluating the regulation of haematopoiesis by discrete niche populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw sequencing data and expression-count data are deposited in GEO, accession number GSE108892. An interactive query and visualization tool for different populations of the bone marrow niche is available at http://aifantislab.com/niche.
Change history
12 July 2019
An Amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Wei, Q. & Frenette, P. S. Niches for hematopoietic stem cells and their progeny. Immunity 48, 632–648 (2018).
Ding, L. & Morrison, S. J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013).
Zhu, J. et al. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood 109, 3706–3712 (2007).
Méndez-Ferrer, S., Lucas, D., Battista, M. & Frenette, P. S. Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 (2008).
Lucas, D. et al. Chemotherapy-induced bone marrow nerve injury impairs hematopoietic regeneration. Nat. Med. 19, 695–703 (2013).
Chow, A. et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 208, 261–271 (2011).
Bruns, I. et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 20, 1315–1320 (2014).
Yamazaki, S. et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 147, 1146–1158 (2011).
Kusumbe, A. P., Ramasamy, S. K. & Adams, R. H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature 507, 323–328 (2014).
Zhou, B. O., Yue, R., Murphy, M. M., Peyer, J. G. & Morrison, S. J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 154–168 (2014).
Mizoguchi, T. et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell 29, 340–349 (2014).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Ramasamy, S. K. Structure and functions of blood vessels and vascular niches in bone. Stem Cells Int. 2017, 5046953 (2017).
Xu, C. et al. Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow. Nat. Commun. 9, 2449 (2018).
Hooper, A. T. et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell 4, 263–274 (2009).
Nombela-Arrieta, C. et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 15, 533–543 (2013).
Crane, G. M., Jeffery, E. & Morrison, S. J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590 (2017).
Ghazanfari, R., Li, H., Zacharaki, D., Lim, H. C. & Scheding, S. Human non-hematopoietic CD271pos/CD140alow/neg bone marrow stroma cells fulfill stringent stem cell criteria in serial transplantations. Stem Cells Dev. 25, 1652–1658 (2016).
Mitroulis, I. et al. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Invest. 127, 3624–3639 (2017).
Yang, G. et al. Osteogenic fate of hypertrophic chondrocytes. Cell Res. 24, 1266–1269 (2014).
Winkler, I. G. et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 18, 1651–1657 (2012).
Cordeiro Gomes, A. et al. Hematopoietic stem cell niches produce lineage-instructive signals to control multipotent progenitor differentiation. Immunity 45, 1219–1231 (2016).
Mrózek, E., Anderson, P. & Caligiuri, M. A. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood 87, 2632–2640 (1996).
Goldman, D. C. et al. BMP4 regulates the hematopoietic stem cell niche. Blood 114, 4393–4401 (2009).
Shi, C. et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating Toll-like receptor ligands. Immunity 34, 590–601 (2011).
Nemeth, M. J., Topol, L., Anderson, S. M., Yang, Y. & Bodine, D. M. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc. Natl Acad. Sci. USA 104, 15436–15441 (2007).
Smith, G. D., Gunnell, D. & Holly, J. Cancer and insulin-like growth factor-I. BMJ 321, 847–848 (2000).
Yu, V. W. et al. Distinctive mesenchymal–parenchymal cell pairings govern B cell differentiation in the bone marrow. Stem Cell Reports 7, 220–235 (2016).
Mauch, P. et al. Hematopoietic stem cell compartment: acute and late effects of radiation therapy and chemotherapy. Int. J. Radiat. Oncol. Biol. Phys. 31, 1319–1339 (1995).
Hérault, A. et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature 544, 53–58 (2017).
Zhou, B. O. et al. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 19, 891–903 (2017).
Van Zant, G. Studies of hematopoietic stem cells spared by 5-fluorouracil. J. Exp. Med. 159, 679–690 (1984).
Klinakis, A. et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473, 230–233 (2011).
Song, R. et al. Mind bomb 1 in the lymphopoietic niches is essential for T and marginal zone B cell development. J. Exp. Med. 205, 2525–2536 (2008).
Poulos, M. G. et al. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Reports 4, 1022–1034 (2013).
Koch, U. et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med. 205, 2515–2523 (2008).
Schmitt, T. M., Ciofani, M., Petrie, H. T. & Zúñiga-Pflücker, J. C. Maintenance of T cell specification and differentiation requires recurrent Notch receptor–ligand interactions. J. Exp. Med. 200, 469–479 (2004).
Lehar, S. M., Dooley, J., Farr, A. G. & Bevan, M. J. Notch ligands Delta1 and Jagged1 transmit distinct signals to T-cell precursors. Blood 105, 1440–1447 (2005).
Olsson, A. et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016).
Paul, F. et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015).
Pronk, C. J. et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1, 428–442 (2007).
Ng, S. Y., Yoshida, T., Zhang, J. & Georgopoulos, K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507 (2009).
Acar, M. et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015).
Pietras, E. M. et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015).
Giladi, A. et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846 (2018).
Jacobsen, S. E. W. & Nerlov, C. Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol. 21, 2–8 (2019).
Passaro, D. et al. CXCR4 is required for leukemia-initiating cell activity in T cell acute lymphoblastic leukemia. Cancer Cell 27, 769–779 (2015).
Pitt, L. A. et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell 27, 755–768 (2015).
Alva, J. A. et al. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn. 235, 759–767 (2006).
DeFalco, J. et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613 (2001).
Kim, J. E., Nakashima, K. & de Crombrugghe, B. Transgenic mice expressing a ligand-inducible Cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. Am. J. Pathol. 165, 1875–1882 (2004).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Economides, A. N. et al. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc. Natl Acad. Sci. USA 110, E3179–E3188 (2013).
Hozumi, K. et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 5, 638–644 (2004).
Lee, E. C. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001).
Reizis, B. & Leder, P. The upstream enhancer is necessary and sufficient for the expression of the pre-T cell receptor α gene in immature T lymphocytes. J. Exp. Med. 194, 979–990 (2001).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Mayer, C. et al. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018).
Rodda, L. B. et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity 48, 1014–1028.e6 (2018).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Acknowledgements
We thank the NYULMC High Performance Computing, Flow Cytometry, Genome Technology Center, Histopathology Core and the Microscopy Laboratory. This research was supported by the US National Institutes of Health (RO1CA202025, RO1CA202027 (I. Aifantis), DK056638, HL069438, DK116312, DK112976 to P.S.F.), the Leukemia & Lymphoma Society (I. Aifantis and A.N.T), the Alex’s Lemonade Stand Foundation for Childhood Cancer (I. Aifantis and A.N.T.), the ERC Advanced grant: European Research Council (AdG 339409, AngioBone) (R.H.A.), the American Cancer Society (RSG-15-189-01-RMC to A.T.) and the St. Baldrick’s Foundation (581357 to A.T.). I. Aifantis thanks the late H. von Boehmer for his support.
Reviewer information
Nature thanks Andreas Trumpp and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.N.T., I.D. and I. Aifantis designed the study and prepared the manuscript. A.N.T. performed the majority of the experiments. I.D. performed all the computational analysis, with guidance in the execution from R.S. and A.T. J.G. generated mouse strains. H.H. and E.H. provided technical assistance with mouse models. M.W. and S.P. performed differentiation assays, with guidance in the execution from P.S.F. All microscopy was performed and interpreted by K.K.S., A.C.-D., M.C.G., A.N.T. and I. Akhmetzyanova, with guidance from R.H.A., D.R.F., J.M.B. and S.K. Y.Z., C.M. and A.H. generated the scRNA-seq data. M.G. and C.D. assisted with transplantation assays. A.E. and R.H.A. provided mouse strains and assisted with data analysis. H.Z. assisted with statistical analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 RNA-seq analysis of the bone marrow microenvironment populations.
a, Representative two-photon imaging of tdTomato+ vascular cells (VE-Cad––tdTomato+), perivascular cells (LEPR––tdTomato+) and osteoblasts (COL2.3––tdTomato+). b, Representative flow cytometry of VE-Cad–tdTomato+, LEPR–tdTomato+ and COL2.3–tdTomato+ populations. c, Principal component analysis of vascular (VE-Cad–tdTomato+, n = 4) (red), perivascular LEPR+ (LEPR–tdTomato+, n = 4) (purple) and osteoblast (COL2.3–tdTomato+, n = 4) (blue) populations, based on the expression of the 1,000 most-variable genes in bulk RNA-seq. d, Relative expression levels of COL2.3+, LEPR+ and VE-Cad+ signature genes across the three subpopulations of the bone marrow niche in bulk RNA-seq. Normalization and statistical analysis were performed using the DESeq2 R package. e, Normalized expression levels of the population-specific markers VE-Cad (Cdh5), LEPR (Lepr) and COL2.3 (Col1a1) for all scRNA-seq clusters. n = 9,622 cells. The data are mean ± s.e.m. Experiments were repeated independently on more than 10 (a, b) biological samples with similar results.
Extended Data Fig. 2 Analysis of VE-Cad+, LEPR+ and COL2.3+ populations.
a, Schematic workflow of independent and integrated analysis of VE-Cad+, LEPR+ and COL2.3+ scRNA-seq data. b, t-SNE representation of VE-Cad+ populations only. Cluster C1 corresponds to arterial cluster V1 (Ly6ahigh). Cluster C2 corresponds to sinusoidal cluster V2 (Stab2high). Cluster C3 is the cycling cluster. c, Normalized expression of arterial, sinusoidal and cycling markers (Ly6a, Stab2 and Mki67, respectively) (n = 4,551 cells). d, Gene signatures of VE-Cad+ subpopulations in the bone marrow, based on the relative expression levels of the ten most-significant markers for each cluster. e, t-SNE representation of the LEPR+ population only. Cluster C1 corresponds to the adipocytic-primed cluster P1 (Mgphigh) and encompasses cluster P2. Cluster C2 corresponds to P3 (Wif1high), and C3 to P4 (Spp1high). f, Normalized expression of P1, P3 and P4 markers (Mgp, Wif1 and Spp1, respectively). n = 3,907 cells. g, Gene signatures of LEPR+ subpopulations in the bone marrow, based on the relative expression levels of the ten most-significant markers for each cluster. h, t-SNE representation of the COL2.3+ population only. Cluster C1 corresponds to cluster O1 (Col16a1high), cluster C2 to O2 (Fbn1high) and C3 to O3 (Bglaphigh). i, Normalized expression of O1, O2 and O3 markers (Col16a1, Fbn1 and Bglap, respectively). n = 1,114 cells. j, Gene signatures of COL2.3+ subpopulations in the bone marrow, based on the relative expression levels of the ten most-significant markers for each cluster. Cluster C4 represents arterial vascular cells (Cdh5, Kdr and Ly6e); C5 is glial-like cells (Fabp7, Mpz and Endrb); and C6 is myocyte-like cells (Pgf, Pln and Acta2). The data shown in c, f, i are mean ± s.e.m. MAST with Bonferroni correction (d, g, j).
Extended Data Fig. 3 Characterization of VE-Cad+ subpopulations.
a, Relative average scRNA-seq expression levels of the previously described arterial and sinusoidal gene signatures at a steady state, within the VE-Cad+ clusters V1 and V2. b, Average scRNA-seq expression levels (left) (n = 4,669 cells) and bone marrow immunofluorescence (right) of arterial expression of SCA-1 (Ly6a), CD102 (Icam2) and PODXL (Podxl) (n = 3 mice); LAMA1 staining (blue) labels all bone vessels; yellow arrowheads indicate arterial vessels. Dashed lines mark bone marrow (bm), compact bone (cb), growth plate (gp) and metaphyseal (mp) bone regions. c, Bone marrow immunofluorescence of arterioles co-stained with SCA-1 and PODXL. n = 3 mice. d, Average scRNA-seq expression levels (left) and bone marrow immunofluorescence (right) of sinusoidal VEGFR3 (Flt4) (red) and CD54 (Icam1) (green) markers. n = 3 mice. LAMA1 staining (blue) labels all bone vessels. e, Average scRNA-seq expression levels and representative flow cytometry analysis of the arterial subpopulation (V1) using SCA-1 and scRNA-seq-identified LY6C (Ly6c1) and CD34 (Cd34) from VE-Cad–tdTomato bone marrow (n = 3 mice). Cells were pre-gated on DAPI−tdTomatohigh cells. The data in b, d, e are mean ± s.e.m.
Extended Data Fig. 4 Characterization of perivascular LEPR+ subpopulations.
a, Relative average scRNA-seq expression levels of the adipocytic- (Hp, Lpl, Adipoq, Slc1a5, Cd302, Gas6 and Apoe) and osteo-associated (Ibsp, Spp1, Alpl, Wif1, Bglap, Sp7 and Runx2) genes, as well as markers used for characterization of LEPR+ cells (Esm1, Vcam1, Cd200 and Cd63). b, ESM1 (green) bone marrow immunofluorescence of LEPR–tdTomato femur. LAMA1 staining (blue) labels all bone vessels. Yellow arrowheads indicate LEPR+ESM1+ cells; white arrowheads indicate LEPR+ESM1− cells. c, CD200 (green) and CD63 (red) bone marrow immunofluorescence of LEPR–tdTomato femur. Nuclei, DAPI (blue). Yellow arrowhead, LEPR+CD200+CD63+ cells; white arrowhead, LEPR+CD200−CD63− cells. d, Human mesenchymal stem cell (hMSC) gene signature module score, overlaid on t-SNE representation. n = 9,622 cells. e, Flow cytometry representation of VCAM1highCD63low and VCAM1lowCD63high cells of tdTomato+ subpopulations, in LEPR–tdTomato bone marrow. Cells were pre-gated on DAPI−tdTomatohigh cells. f, Fibroblastic colony-forming unit activity of sorted total LEPR+ (purple), LEPR+VCAM1lowCD63high (maroon) and LEPR+VCAM1highCD63low (yellow) cells from bone marrow of LEPR–tdTomato mice (n = 8). The data are mean ± s.d. (f). N.S., not significant, *P ≤ 0.05, **P ≤ 0.01. Student’s t-test, two-tailed (f). Data are representative of two (b, c) or three (e, f) independent experiments.
Extended Data Fig. 5 Characterization of COL2.3+ subpopulations and cycling cells.
a, Relative average scRNA-seq expression levels of O1- (Edil3, Mmp14, Ostn, Col12a1, Angptl2 and Col16a1), O2- (Sox9, Comp, Chad and Col10a1) and O3- (Col11a2, Col1a2, Sparc, Bglap2 and Bglap3) associated genes. b–d, Average scRNA-seq expression levels (n = 9,622 cells) and bone marrow immunofluorescence of MMP14 (green) (b) (n = 3 mice), CD9 (green) (c) (n = 3 mice) and CAR3 (green) (d) (n = 3 mice) in COL2.3–tdTomato femur, with arrows indicating co-staining with tdTomato (red). Nuclei, DAPI (blue). Arrowhead, COL2.3+MMP14+ (b), COL2.3+CD9+ (c) and COL2.3+CAR3+ (d). e, Expression levels of Mki67 in all identified subpopulations. n = 9,622 cells. f, Enriched Gene Ontology biological processes terms that are most-strongly associated with cycling cluster (C), colour-coded by the significance of enrichment and size on the basis of the fraction of overlapping genes. n = 9,622 cells. g, Contribution of VE-Cad–tdTomato+, LEPR–tdTomato+ and COL2.3–tdTomato+ cells to the cycling cluster at a steady state (n = 70 cells). The data in b–e are mean ± s.e.m.
Extended Data Fig. 6 Effect of treatment with 5-FU on subsets of the bone marrow niche.
a, Representative haematoxylin and eosin-stained sections of bone marrow on day five after treatment with control (PBS) or 5-FU (n = 3 mice). b, Frequency and numbers of bone marrow LSK cells on day five after treatment with control (CNTRL) (n = 4) or 5-FU (n = 5). c, Absolute numbers of bone marrow niche cells, vascular VE-Cad+ (control, n = 4; 5-FU, n = 10), perivascular LEPR+ (control, n = 2; 5-FU, n = 5) and COL2.3+ osteoblasts (control, n = 4; 5-FU, n = 5) from mice treated with PBS or 5-FU. d, Gene signatures of LEPR+ subpopulations (including cluster P5) on the basis of the average relative expression levels of the ten most-significant markers for each cluster, exclusively within the LEPR+ subset. MAST with Bonferroni correction. e, Relative expression levels of upregulated adipogenesis-associated genes and downregulated osteogenesis-associated genes in LEPR+ subpopulations in response to treatment with 5-FU. f, Pathways enriched in LEPR+ cells in response to treatment with 5-FU (n = 17,374 cells). Fisher’s exact test. g, Contribution of VE-Cad–tdTomato+, LEPR–tdTomato+ and COL2.3–tdTomato+ cells to the cycling cluster after treatment with 5-FU (n = 418 cells). h, Expression levels of Mki67 in all identified subpopulations at a steady state, and after treatment with 5-FU (n = 17,374 cells). The data are mean ± s.d. N.S., not significant, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Student’s t-test, two-tailed (b, c). The data in h are mean ± s.e.m.
Extended Data Fig. 7 Validation of scRNA-seq in VE-Cad+ and LEPR+ cells following treatment.
a, b, Heat map and hierarchical clustering of mean normalized expression values of VE-Cad+ (a) and LEPR+ (b) control-treated and 5-FU-treated samples for 5-FU-modulated genes (top 50 differentially expressed) in two independent scRNA-seq experiments. c, d, log-transformed fold changes of differentially expressed genes (up- and downregulated > 1.2×, adjusted P value < 0.001) in 5-FU-treated versus control-treated VE-Cad+ (n = 697 genes) cells (c) and LEPR+ (n = 829 genes) cells (d), for two independent experiments. The trend line (dashed) and the confidence interval (grey shading) were calculated using the linear model. MAST with Bonferroni correction. VE-Cad+ control-treated, n = 5,796 cells; VE-Cad+ 5-FU-treated, n = 1,481 cells; LEPR+ control-treated, n = 6,128 cells; LEPR+ 5-FU-treated, n = 4,867 cells.
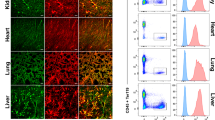
Extended Data Fig. 8 Analysis of Dll4-mCherry, Dll1-mCherry and Jag1-mCherry reporter mice.
a, Reverse-transcription PCR of Dll4, Dll1 and Jag1 in vascular cells (red), perivascular LEPR+ cells (purple) and osteoblasts (blue), normalized to Gapdh (n = 4 mice). b, Low-magnification immunofluorescence images of thymus sections from Dll4-mCherry, Dll1-mCherry and Jag1-mCherry mice. c, Representative flow cytometry, measuring mCherry fluorescence in total bone marrow of Dll4-mCherry, Dll1-mCherry and JAG1-mCherry mice. Indicated values represent percentages of the complete CD144+ and mCherry+CD144− populations. Cells were pre-gated on DAPI− cells. d, Representative mCherry levels in DAPI−CD45lowTER119lowCD144+ bone marrow endothelial cells from Dll4-mCherry (red) (n = 4), Dll1-mCherry (blue) (n = 3), Jag1-mCherry (black) (n = 4) and control (grey) (n = 5) mice. e, f, Representative immunofluorescence metaphysis and diaphysis of Dll4-mCherry (e) and Dll1-mCherry (f) bone marrow (n = 3 mice). mCherry (red) and LAMA1 (blue). g, h, Representative two-photon images of bone marrow from intact (left) or dextran-injected (right) Dll4-mCherry (g) and Dll1-mCherry (h) mice (n = 3 mice). i, Normalized counts of key differentially expressed genes from bulk RNA-seq performed on CD144−DLL1+ cells (purple) (from n = 2 mice) and CD144+DLL1+ cells (black) (from n = 2 mice). j, Representative flow cytometry histogram measuring mCherry fluorescence in NK1.1+ population from Dll1-mCherry (pink) and control (black) mice (n = 3 mice). The data are mean ± s.d. N.S., not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t-test, two-tailed. Data are representative of two (a, e–h, j) or three (b–d) independent experiments.
Extended Data Fig. 9 Gene expression program of myeloid differentiation is enhanced in VE-Cad-Dll4i3COIN HSPCs.
a, b, Representative percentage of bone marrow progenitors in VE-CadcreER-Dll4i3COIN and littermate-control mice for common lymphoid progenitors gate (CLP gate) (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 11) (a) and granulocyte–monocyte progenitors gate (GMP gate) (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 13) (b). c, Frequencies of CD3+ T cells (control, n = 12; VE-CadcreER-Dll4i3COIN, n = 11). d, Total numbers of mature haematopoietic subsets in tamoxifen-treated VE-CadcreER-Dll4i3COIN and littermate-control mice, including B220+ B cells (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 10), CD3+ T cells (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 10) and CD11b+GR1+ myeloid cells (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 10). e, Absolute numbers of thymocytes from VE-CadcreER-Dll4i3COIN mice (n = 11) and littermate-control mice (n = 9). f, Representative flow cytometry analysis of thymic subsets in tamoxifen-treated VE-CadcreER-Dll4i3COIN and littermate-control mice. g, Frequencies (control, n = 8; VE-CadcreER-Dll4i3COIN, n = 5) and absolute numbers (control, n = 6; VE-CadcreER-Dll4i3COIN, n = 4) of early thymic progenitors in thymi from VE-CadcreER-Dll4i3COIN and littermate-control mice. h, Percentage (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 10) of HSCs, MPP2 cells, MPP3–4 cells, MPP4 cells and LSK cells from the bone marrow of VE-CadcreER-Dll4i3COIN and littermate-control mice. i, Total numbers of bone marrow HSCs, MPP2 cells, MPP3 cells, MPP4 cells and LSK cells from VE-CadcreER-Dll4i3COIN and littermate-control mice (control, n = 10; VE-CadcreER-Dll4i3COIN, n = 10). j, Representative immunofluorescence of early progenitors (Lin−CD48−CD150+) adjacent to the DLL4-producing vascular endothelium in Dll4-mCherry. Lin cocktail, CD11b, GR1, CD41, TER119 and B220. Arrowhead, Lin−CD150+ progenitors. n = 3 mice. k, scRNA-seq t-SNE visualization of the LSK compartment (n = 21,116 cells), colour-coded by genotype. l, m, Distribution of enrichment scores for myeloid progenitor (l) and HSC (m) gene signatures within the scRNA-seq-profiled HSPC populations from bone marrow of tamoxifen-treated VE-CadcreER-Dll4i3COIN mice (pooled n = 2) and littermate-control mice (pooled n = 2). n = 21,116 cells. The data are mean ± s.d. NS, not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t-test, two-tailed (a–d, g–i) or Wilcoxon rank-sum test (l, m). Data are representative of four (a–e) or two (g) independent experiments.
Extended Data Fig. 10 Deletion of endothelial Dll1 does not affect early lineage priming of haematopoietic progenitors.
a–f, Flow cytometry analysis of bone marrow progenitors in VE-Cad-Dll1fl/fl and littermate-control mice, showing equivalent frequencies of common lymphoid progenitors (control, n = 10; VE-Cad-Dll1fl/fl, n = 8) (a), granulocyte–monocyte progenitors (control, n = 7; VE-Cad-Dll1fl/fl, n = 9) (b) and MPP4 cells (control, n = 10; VE-Cad-Dll1fl/fl, n = 10) (c), of B220+ B cells (control, n = 10; VE-Cad-Dll1fl/fl, n = 10) (d, f), CD3+ T cells (control, n = 10; VE-Cad-Dll1fl/fl, n = 10) (d, f) and CD11b+GR1+ monocytic–granulocytic subset (control, n = 8; VE-Cad-Dll1fl/fl, n = 9) (e, f). g, Absolute numbers of thymocytes from VE-Cad-Dll1fl/fl and littermate-control mice (control, n = 6; VE-Cad-Dll1fl/fl, n = 10). h, Representative flow cytometry analysis of thymic subsets in VE-Cad-Dll1fl/fl and littermate-control mice. i, Frequencies (control, n = 4; VE-Cad-Dll1fl/fl, n = 6) and absolute numbers (control, n = 4; VE-Cad-Dll1fl/fl, n = 6) of early thymic progenitors from thymi of VE-Cad-Dll1fl/fl and littermate-control mice. The data are mean ± s.d. N.S., not significant, Student’s t-test, two-tailed. Data are representative of three independent experiments.
Supplementary information
Supplementary Information
This file contains a full guide for Supplementary Tables 1–5.
Supplementary Tables
This file contains Supplementary Tables 1–5. Full table legends appear in a separate PDF file.
Source data
Rights and permissions
About this article
Cite this article
Tikhonova, A.N., Dolgalev, I., Hu, H. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019). https://doi.org/10.1038/s41586-019-1104-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1104-8
This article is cited by
-
SOXC are critical regulators of adult bone mass
Nature Communications (2024)
-
Profiling joint tissues at single-cell resolution: advances and insights
Nature Reviews Rheumatology (2024)
-
Clonal hematopoiesis and its impact on the aging osteo-hematopoietic niche
Leukemia (2024)
-
Generating human bone marrow organoids for disease modeling and drug discovery
Nature Protocols (2024)
-
Oncostatin M: Dual Regulator of the Skeletal and Hematopoietic Systems
Current Osteoporosis Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.