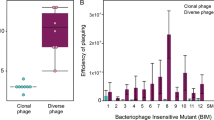
Abstract
About half of all bacteria carry genes for CRISPR–Cas adaptive immune systems1, which provide immunological memory by inserting short DNA sequences from phage and other parasitic DNA elements into CRISPR loci on the host genome2. Whereas CRISPR loci evolve rapidly in natural environments3,4, bacterial species typically evolve phage resistance by the mutation or loss of phage receptors under laboratory conditions5,6. Here we report how this discrepancy may in part be explained by differences in the biotic complexity of in vitro and natural environments7,8. Specifically, by using the opportunistic pathogen Pseudomonas aeruginosa and its phage DMS3vir, we show that coexistence with other human pathogens amplifies the fitness trade-offs associated with the mutation of phage receptors, and therefore tips the balance in favour of the evolution of CRISPR-based resistance. We also demonstrate that this has important knock-on effects for the virulence of P. aeruginosa, which became attenuated only if the bacteria evolved surface-based resistance. Our data reveal that the biotic complexity of microbial communities in natural environments is an important driver of the evolution of CRISPR–Cas adaptive immunity, with key implications for bacterial fitness and virulence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data used in this study are available on figshare at https://doi.org/10.6084/m9.figshare.9752903.
References
Grissa, I., Vergnaud, G. & Pourcel, C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36, W145–W148 (2008).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Andersson, A. F. & Banfield, J. F. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320, 1047–1050 (2008).
Laanto, E., Hoikkala, V., Ravantti, J. & Sundberg, L. R. Long-term genomic coevolution of host–parasite interaction in the natural environment. Nat. Commun. 8, 111 (2017).
Westra, E. R. et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049 (2015).
van Houte, S., Buckling, A. & Westra, E. R. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 80, 745–763 (2016).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
O’Toole, G. A. Cystic fibrosis airway microbiome: overturning the old, opening the way for the new. J. Bacteriol. 200, 1–8 (2018).
Folkesson, A. et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851 (2012).
Roach, D. R. & Debarbieux, L. Phage therapy: awakening a sleeping giant. Emerg. Top. Life Sci. 1, 93–103 (2017).
Rossitto, M., Fiscarelli, E. V. & Rosati, P. Challenges and promises for planning future clinical research into bacteriophage therapy against Pseudomonas aeruginosa in cystic fibrosis. An argumentative review. Front. Microbiol. 9, 775 (2018).
De Smet, J., Hendrix, H., Blasdel, B. G., Danis-Wlodarczyk, K. & Lavigne, R. Pseudomonas predators: understanding and exploiting phage–host interactions. Nat. Rev. Microbiol. 15, 517–530 (2017).
Chabas, H., van Houte, S., Høyland-Kroghsbo, N. M., Buckling, A. & Westra, E. R. Immigration of susceptible hosts triggers the evolution of alternative parasite defence strategies. Proc. R. Soc. B 283, 20160721 (2016).
Harrison, F. Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923 (2007).
O’Brien, S. & Fothergill, J. L. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 364, 1–10 (2017).
Bhargava, N., Sharma, P. & Capalash, N. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect. Immun. 82, 3417–3425 (2014).
Rocha, G. A. et al. Species distribution, sequence types and antimicrobial resistance of Acinetobacter spp. from cystic fibrosis patients. Epidemiol. Infect. 146, 524–530 (2018).
Diraviam Dinesh, S. & Diraviam Dinesh, S. Artificial sputum medium. Protoc. Exchange https://doi.org/10.1038/protex.2010.212 (2010).
An, D., Danhorn, T., Fuqua, C. & Parsek, M. R. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl Acad. Sci. USA 103, 3828–3833 (2006).
León, M. & Bastías, R. Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 6, 343 (2015).
Kavanagh, K. & Reeves, E. P. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28, 101–112 (2004).
Hernandez, R. J. et al. Using the wax moth larva Galleria mellonella infection model to detect emerging bacterial pathogens. PeerJ 6, e6150 (2019).
Craig, L., Pique, M. E. & Tainer, J. A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 (2004).
Johnson, P. T. J., de Roode, J. C. & Fenton, A. Why infectious disease research needs community ecology. Science 349, 1259504 (2015).
Alizon, S., de Roode, J. C. & Michalakis, Y. Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567 (2013).
Benmayor, R., Hodgson, D. J., Perron, G. G. & Buckling, A. Host mixing and disease emergence. Curr. Biol. 19, 764–767 (2009).
Keesing, F. et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652 (2010).
Chabas, H. et al. Evolutionary emergence of infectious diseases in heterogeneous host populations. PLoS Biol. 16, e2006738 (2018).
van Houte, S. et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016).
Wright, R. C. T., Friman, V. P., Smith, M. C. M. & Brockhurst, M. A. Cross-resistance is modular in bacteria–phage interactions. PLoS Biol. 16, e2006057 (2018).
Martínez-García, E., Calles, B., Arévalo-Rodríguez, M. & de Lorenzo, V. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 11, 38 (2011).
Goto, M. et al. Real-time PCR method for quantification of Staphylococcus aureus in milk. J. Food Prot. 70, 90–96 (2007).
R Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (R Foundation for Statistical Computing, 2018).
Wickham, H. tidyverse: Easily Install and Load the 'Tidyverse'. R package version 1.2.1 https://cran.r-project.org/web/packages/tidyverse/index.html (2017).
Therneau, T. A Package for Survival Analysis in S. R package version 2.38 https://CRAN.R-project.org/package=survival (2015).
Acknowledgements
We thank A. Buckling for critical reading of the manuscript, J. Common, E. Hesse and S. Meaden for comments on the manuscript, and J. P. Pirnay and D. de Vos for sharing clinical isolates of S. aureus, A. baumannii and B. cenocepacia. This work was supported by grants from the ERC (ERC-STG-2016-714478 - EVOIMMECH) and the NERC (NE/M018350/1), which were awarded to E.R.W.
Author information
Authors and Affiliations
Contributions
Conceptualization of the study was done by E.O.A. and E.R.W. Experimental design was carried out by E.O.A., A.M.L., C.R. and E.R.W. Adsorption and infection assays were done by E.O.A. All evolution experiments were performed by E.O.A., E.P. and I.M. E.O.A. performed the DNA extractions and qPCR reactions, and the competition experiments, virulence assays and motility assays were performed by E.O.A. and E.P. Formal analysis of results was done by E.O.A., E.P., C.R. and E.R.W. The original draft was written by E.O.A., with later edits and reviews by E.O.A. and E.R.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Only P. aeruginosa adsorbs phage DMS3vir.
Phage levels (in p.f.u. ml−1) in minutes after infection of P. aeruginosa PA14 and three other bacterial species (n = 84 biologically independent replicates). Controls were carried out in the absence of bacteria. Here, the lines are regression slopes with shaded areas corresponding to 95% confidence intervals. Linear model: effect of P. aeruginosa on phage titre over time; t = −3.37, P = 0.0009; S. aureus; t = 1.63, P = 0.11; A. baumannii; t = 1.20, P = 0.23; B. cenocepacia; t = −0.27, P = 0.79; overall model fit; F9,235 = 4.33, adjusted R2 = 0.11, P = 3.17 × 10−5
Extended Data Fig. 2 Enhanced CRISPR resistance evolution in ASM.
Proportion of P. aeruginosa that acquired surface modification or CRISPR-based immunity (or remained sensitive) 3 d.p.i. with phage DMS3vir when grown in ASM (6 replicates per treatment, with 24 colonies screened from each replicate, n = 30 biologically independent replicates). Deviance test: relationship between community composition and CRISPR; residual deviance (25, n = 30) = 1.26, P = 2.2 × 10−16; Tukey contrasts: monoculture versus mixed; z = −5.30, P = 1 × 10−4; monoculture versus A. baumannii; z = −5.60, P = 1 × 10−4; monoculture versus B. cenocepacia; z = −2.80, P = 0.02; monoculture versus S. aureus; z = −0.76, P = 0.93. Data are mean ± s.e.m
Extended Data Fig. 3 Increased evolution of CRISPR-based resistance across a range of microbial community compositions over time.
Proportion of P. aeruginosa that acquired surface modification or CRISPR-based immunity (or remained sensitive) at up to 3 d.p.i. with phage DMS3vir when grown either in monoculture (100%) or in polyculture mixtures consisting of the mixed microbial community but with varying starting percentages of P. aeruginosa based on volume (6 replicates for most samples, with 24 colonies per replicate, n = 42 biologically independent replicates for a, n = 32 biologically independent replicates for b, and n = 42 biologically independent replicates for c). a, Resistance evolution at 1 d.p.i. Data are mean ± s.e.m. Deviance test: relationship between CRISPR and P. aeruginosa starting percentage at time point 1; residual deviance (35, n = 42) = 4.42, P = 0.004; 1%; z = −3.27, P = 0.002; 10%; z = 1.21, P = 0.23; 25%; z = 1.62, P = 0.11; 50%; z = 2.20, P = 0.034; 90%; z = 2.07, P = 0.046; 99%; z = 0.47, P = 0.65; 100%; z = 1.47, P = 0.15. b, Resistance evolution at 2 d.p.i. Data are mean ± s.e.m. Deviance test: relationship between CRISPR and P. aeruginosa starting percentage at time point 2; residual deviance (25, n = 32) = 3.86, P = 2.51 × 10−6; 1%; z = −2.14, P = 0.04; 10%; z = 1.19, P = 0.25; 25%; z = 2.07, P = 0.049; 50%; z = 1.89, P = 0.07; 90%; z = 1.12, P = 0.27; 99%; z = 1.21, P = 0.24; 100%; z = 1.11, P = 0.28. c, Resistance evolution at 3 d.p.i. Data are mean ± s.e.m. Deviance test: relationship between CRISPR and P. aeruginosa starting percentage at time point 3; residual deviance (35, n = 42) = 8.24, P = 0.0004; 1%; z = −3.38, P = 0.002; 10%; z = 2.12, P = 0.04; 25%; z = 2.77, P = 0.009; 50%; z = 3.07, P = 0.004; 90%; z = 2.46, P = 0.019; 99%; z = 1.55, P = 0.13; 100%; z = 0.87, P = 0.39
Extended Data Fig. 4 Microbial community composition affects phage epidemic size.
The DMS3vir phage titres (in p.f.u. ml−1) over time up to 3 d.p.i. of P. aeruginosa grown either in monoculture (100%) or in polyculture mixtures as shown in Extended Data Fig. 3. Each data point represents the mean, error bars denote s.e.m. (n = 171 independent biological samples). Two-way ANOVA: overall effect of P. aeruginosa starting percentage on phage titre; F6,105 = 14.84, P = 1.1 × 10−12
Extended Data Fig. 5 No correlation between phage epidemic size and evolution of CRISPR resistance.
The correlation between the proportion of evolved phage-resistant clones with CRISPR-based resistance and the phage epidemic sizes (in p.f.u. ml−1) in the presence of other bacterial species, using data taken from experiments shown in Fig. 1, Extended Data Figs. 2, 3c and 6 (n = 137 biologically independent samples per time point). Correlations are separated by day, as phage titres were measured daily. Here, the lines are regression slopes, with shaded areas corresponding to 95% confidence intervals. Pearson’s product–moment correlation tests between phage titres (at each day after infection) and levels of CRISPR-based resistance: T = 1; t136 = −0.02, P = 0.98, R2 = −0.002; T = 2; t136 = 0.59, P = 0.55, R2 = 0.05; T = 3; t136 = −0.90, P = 0.37, R2 = −0.08
Extended Data Fig. 6 Starting phage titre does not affect CRISPR evolution in the presence of a microbial community.
Proportion of P. aeruginosa that acquired CRISPR-based resistance at 3 d.p.i. with varying starting titres of phage DMS3vir when grown in polyculture (6 replicates per treatment, with 24 colonies per replicate, n = 24 biologically independent replicates). Deviance test: start phage and CRISPR; residual deviance (20, n = 24) = 2.00, P = 0.13; Tukey contrasts: 102 versus 104; z = −1.52, P = 0.42; 104 versus 106; z = −0.76, P = 0.87; 106 versus 108; z = 1.31, P = 0.56; 102 versus 106; z = −2.24, P = 0.11; 102 versus 108; z = −0.99, P = 0.75; 104 versus 108; z = 0.56, P = 0.94. Data are mean ± s.e.m
Extended Data Fig. 7 LPS-based phage resistance also affects in vivo virulence.
Time until death (given as median ± one standard error) for G. mellonella larvae infected with PA14 clones that evolved phage resistance by LPS modification, compared to the phage-sensitive ancestral (n = 209 biologically independent samples). Cox proportional hazards model with Tukey contrasts: sensitive (ancestral) versus LPS; z = 4.81, P = 1.49 × 10−6. overall model fit; LRT3 = 44.94, P = 1 × 10−9
Supplementary information
Rights and permissions
About this article
Cite this article
Alseth, E.O., Pursey, E., Luján, A.M. et al. Bacterial biodiversity drives the evolution of CRISPR-based phage resistance. Nature 574, 549–552 (2019). https://doi.org/10.1038/s41586-019-1662-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1662-9
This article is cited by
-
CRISPR-Cas provides limited phage immunity to a prevalent gut bacterium in gnotobiotic mice
The ISME Journal (2023)
-
The highly diverse antiphage defence systems of bacteria
Nature Reviews Microbiology (2023)
-
Interspecific competition can drive plasmid loss from a focal species in a microbial community
The ISME Journal (2023)
-
Bacterial volatile organic compounds attenuate pathogen virulence via evolutionary trade-offs
The ISME Journal (2023)
-
Mucin induces CRISPR-Cas defense in an opportunistic pathogen
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.