Abstract
The current need for novel antibiotics is especially acute for drug-resistant Gram-negative pathogens1,2. These microorganisms have a highly restrictive permeability barrier, which limits the penetration of most compounds3,4. As a result, the last class of antibiotics that acted against Gram-negative bacteria was developed in the 1960s2. We reason that useful compounds can be found in bacteria that share similar requirements for antibiotics with humans, and focus on Photorhabdus symbionts of entomopathogenic nematode microbiomes. Here we report a new antibiotic that we name darobactin, which was obtained using a screen of Photorhabdus isolates. Darobactin is coded by a silent operon with little production under laboratory conditions, and is ribosomally synthesized. Darobactin has an unusual structure with two fused rings that form post-translationally. The compound is active against important Gram-negative pathogens both in vitro and in animal models of infection. Mutants that are resistant to darobactin map to BamA, an essential chaperone and translocator that folds outer membrane proteins. Our study suggests that bacterial symbionts of animals contain antibiotics that are particularly suitable for development into therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information or have been deposited to the indicated databases. The genome of P. khanii HGB1456 has been deposited to GenBank with accession number WHZZ00000000. The transcriptomic dataset (Extended Data Fig. 7) has been deposited to NCBI Sequence Read Archive with identifier PRJNA530781. The mass spectrometry proteomics (Extended Data Fig. 8 and Supplementary Table 2) data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013319. Source Data for Figs. 2c, 4 and Extended Data Figs. 5b, 9 are provided with the paper. All other data are available from the corresponding author.
Change history
25 March 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387 (2013).
Lomovskaya, O. & Lewis, K. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl Acad. Sci. USA 89, 8938–8942 (1992).
Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204 (2004).
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018).
Brown, E. D. & Wright, G. D. Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016).
Crawford, J. M. & Clardy, J. Bacterial symbionts and natural products. Chem. Commun. 47, 7559–7566 (2011).
Tobias, N. J., Shi, Y. M. & Bode, H. B. Refining the natural product repertoire in entomopathogenic bacteria. Trends Microbiol. 26, 833–840 (2018).
Tambong, J. T. Phylogeny of bacteria isolated from Rhabditis sp. (Nematoda) and identification of novel entomopathogenic Serratia marcescens strains. Curr. Microbiol. 66, 138–144 (2013).
Yokoyama, K. & Lilla, E. A. C–C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 35, 660–694 (2018).
Schramma, K. R., Bushin, L. B. & Seyedsayamdost, M. R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat. Chem. 7, 431–437 (2015).
Embley, T. M. & Stackebrandt, E. The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 48, 257–289 (1994).
Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome Med. 8, 51 (2016).
Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 (2016).
O’Shea, R. & Moser, H. E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 51, 2871–2878 (2008).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Konovalova, A., Kahne, D. E. & Silhavy, T. J. Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 (2017).
Ghequire, M. G. K., Swings, T., Michiels, J., Buchanan, S. K. & De Mot, R. Hitting with a BAM: selective killing by lectin-like bacteriocins. mBio 9, e02138-17 (2018).
Storek, K. M. et al. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl Acad. Sci. USA 115, 3692–3697 (2018).
Hart, E. M. et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl Acad. Sci. USA 116, 21748–21757 (2019).
Hartmann, J.-B., Zahn, M., Burmann, I. M., Bibow, S. & Hiller, S. Sequence-specific solution NMR assignments of the β-barrel insertase BamA to monitor its conformational ensemble at the atomic level. J. Am. Chem. Soc. 140, 11252–11260 (2018).
Kaur, H. et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR 73, 375–384 (2019).
Ramos-Castañeda, J. A. et al. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: systematic review and meta-analysis: mortality due to KPC Klebsiella pneumoniae infections. J. Infect. 76, 438–448 (2018).
Xu, L., Sun, X. & Ma, X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16, 18 (2017).
Sun, J., Zhang, H., Liu, Y. H. & Feng, Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808 (2018).
Levasseur, P. et al. Efficacy of a ceftazidime–avibactam combination in a murine model of septicemia caused by Enterobacteriaceae species producing ampc or extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 58, 6490–6495 (2014).
Wunderink, R. G. et al. Effect and safety of meropenem–vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II randomized clinical trial. Infect. Dis. Ther. 7, 439–455 (2018).
King, A. M. et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 510, 503–506 (2014).
Liu, J., Smith, P. A., Steed, D. B. & Romesberg, F. Efforts toward broadening the spectrum of arylomycin antibiotic activity. Bioorg. Med. Chem. Lett. 23, 5654–5659 (2013).
Smith, P. A. et al. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 561, 189–194 (2018).
Richter, M. F. et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304 (2017).
Crits-Christoph, A., Diamond, S., Butterfield, C. N., Thomas, B. C. & Banfield, J. F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444 (2018).
Tobias, N. J. et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2, 1676–1685 (2017).
Pantel, L. et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell 70, 83–94.e7 (2018).
Racine, E. et al. In vitro and in vivo characterization of NOSO-502, a novel inhibitor of bacterial translation. Antimicrob. Agents Chemother. 62, e01016–e01018 (2018).
Poinar, G. Jr. Origins and phylogenetic relationships of the entomophilic rhabditids, Heterorhabditis and Steinernema. Fundam. Appl. Nematol. 16, 333–338 (1993).
Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 351, 180–186 (2016).
Antipov, D., Korobeynikov, A., McLean, J. S. & Pevzner, P. A. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32, 1009–1015 (2016).
Blin, K. et al. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41 (2017).
Lassak, J., Henche, A. L., Binnenkade, L. & Thormann, K. M. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 76, 3263–3274 (2010).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Tang, X. et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 10, 2841–2849 (2015).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Murphy, K. C. & Campellone, K. G. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4, 11 (2003).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Mateus, A. et al. Thermal proteome profiling in bacteria: probing protein state in vivo. Mol. Syst. Biol. 14, e8242 (2018).
Becher, I. et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 12, 908–910 (2016).
Hughes, C. S. et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757 (2014).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019).
Sridharan, S. et al. Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat. Commun. 10, 1155 (2019).
Franken, H. et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 10, 1567–1593 (2015).
Alvarez, F. J. D., Orelle, C. & Davidson, A. L. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 132, 9513–9515 (2010).
Ritchie, T. K. et al. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Roman-Hernandez, G., Peterson, J. H. & Bernstein, H. D. Reconstitution of bacterial autotransporter assembly using purified components. eLife 3, e04234 (2014).
Hagan, C. L., Kim, S. & Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 (2010).
Acknowledgements
This work was supported by NIH grant P01 AI118687 to K.L. and K.E.N. A. Mateus was supported by a fellowship from the EMBL Interdisciplinary Postdoc (EI3POD) Programme under Marie Skłodowska-Curie Actions COFUND (grant number 664726). S.H. was supported by the Swiss National Science Foundation via the NFP 72 (407240_167125). N.N. was supported by NIH grants GM127896 and GM127884. We thank H. Goodrich-Blair for providing strains of Photorhabdus and Xenorhabdus; M. Kagan for help with isolating darobactin; the Northeastern University Barnett Institute MS Core Facility for access to its LC–MS resources; D. Baldisseri from Bruker Biospin Corporation for recording some of the NMR data of darobactin; N. Kurzawa for the help with the analysis of thermal proteome profiling data; W. Fowle for assistance with scanning electron microscopy experiments; Y. Su for assistance with the ITC experiments; and R. Machado for help with taxonomy of Photorhabdus.
Author information
Authors and Affiliations
Contributions
K.L. designed the study, analysed results and wrote the paper. Y.I. identified darobactin, designed the study and analysed results. K.J.M. designed the animal study, wrote the paper and analysed results. A.I. performed mass spectrometry and, with M.M., isolated darobactin. Q.F.-G., C.H., X.M., J.J.G. and A. Makriyannis identified the structure of darobactin. A.D. provided logistical support for the study. S.M. performed microscopy studies and analysed data. M.C. and M.G. performed susceptibility studies. S.N. performed animal studies. T.F.S., R.G., N.B., Z.G.W. and L.L.-O. identified darobactin BGCs and generated the knockout and heterologous expression strains. H.K. performed the NMR studies of BamA. S.H. designed and analysed the NMR studies and wrote the paper. R.W. performed the BAM nanodisc studies. N.N. designed and analysed the BAM nanodisc studies and wrote the paper. A.T., M.M.S. and A. Mateus performed the proteomics study and analysed data. K.E.N., J.L.E. and A.O. performed the transcriptome study and analysed data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Eric Brown, Tilmann Weber and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Structural determination of darobactin.
a, HPLC chromatogram of darobactin. Inset, high-resolution mass spectra (HRMS) of darobactin showing a peak at m/z 966.41047, which corresponds to the [M + H]+ ion and another at m/z 483.70865, which corresponds to [M + 2H]2+ ion. b, Higher-energy collisional dissociation–MS/MS spectra of darobactin. c, 1H NMR spectrum of darobactin. d, 13C NMR spectrum. e, 1H-13C HMBC NMR spectrum. f, 1H-13C HSQC NMR spectrum. g, COSY NMR spectrum. h, ROESY NMR spectrum.
Extended Data Fig. 2 NMR assignments of darobactin.
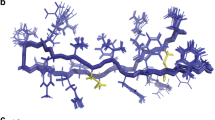
a, 1H, 13C and 15N NMR chemical shifts (ppm) for darobactin. †Owing to overlap with a residual water peak at 4.6 ppm, the multiplicity and J coupling values were from a different 1H NMR spectrum of darobactin in water:deuterated acetonitrile (2:1, v/v). ‡Two partially overlapping peaks were observed at 131.79 ppm and 131.83 ppm. b, Structure of darobactin with numbering for NMR assignments. c, Key ROESY correlations (top) and three-dimensional model of darobactin (bottom).
Extended Data Fig. 3 BGC of darobactin in selected bacterial strains.
a, The BGC consists of the structural gene darA (coloured in blue), darBCD (transporter encoding genes; grey) and darE (a radical SAM enzyme; orange). In addition, a relE-like gene (black) open-reading frame is co-located with the BGC at different positions in different species. The BGC can be detected in most Photorhabdus strains in a conserved genetic region. In addition, homologous BGCs (related genes show the identical colour code) can be found in Yersinia, Vibrio and Pseudoalteromonas strains. b, Biosynthetic hypothesis. The propeptide encoded by darA consists of 58 amino acids. The crosslinks are installed on the linear propeptide by DarE. In a next step, the leader and tail regions are cleaved off and darobactin is secreted by the ABC transporter DarBCD. c, The amino acid sequence of the propeptide from selected bacterial strains. The darobactin core peptide is highlighted in bold and the amino acids involved in the crosslinking in bold red. The asterisk indicates the stop codon.
Extended Data Fig. 4 Darobactin knockout strain and heterologous expression, and putative structures and producers of darobactin A–E.
a, Schematic of the double crossover knockout vector pNB02 and the targeted genomic region. b, Schematic of the darobactin BGC expression plasmid. c, Test PCRs on P. khanii DSM3369 ΔdarABCDE, showing the loss of the darobactin BGC. Left, amplification of darA (primers darA_f/r) results in a 177-bp fragment in the wild-type (WT) strain and in no fragment in the mutant. Right, after loss of pNB02 (indicated by sensitivity to kanamycin), amplification of a 450-bp fragment if the BGC is deleted (primers DSMko_f/r) occurs. Positive controls include pNB03-darA-E and pNB02. Primer positions are indicated in blue in a. The raw DNA gel is provided in Supplementary Fig. 1. d, LC–MS-extracted ion chromatogram at m/z = 483.7089 ± 0.001. Yellow, P. khanii DSM3369 ΔdarABCDE and pNB03; red, P. khanii DSM3369 ΔdarABCDE and pNB03-darA-E; brown, E. coli BW25113 and pNB03-darA-E; blue, P. khanii DSM3369 wild type. Inset, HRMS spectrum of the ion peak showing the double charged [M + 2H]2+ ion that corresponds to darobactin. c, d, Data are representative of at least three independent biological replicates. e, Putative darobactin analogues B–E were drawn based on the amino acid sequence that is present in the darobactin BGC. The proposed darobactin-producing organisms were identified by a BLASTp search of the seven-amino-acid sequence of darobactin A, confirming the presence of darBCDE downstream of the propeptide. Amino acid changes from darobactin A are highlighted in red. f, The propeptide sequence of the various darobactin analogues.
Extended Data Fig. 5 Darobactin mechanism of action and resistance studies.
a, Darobactin and polymyxin B MIC studies against E. coli MG1655 were performed in the presence of LPS. Addition of LPS antagonized polymyxin activity, but not darobactin activity. Data are mean ± s.d. of triplicate experiments. b, Groups of five mice were infected intraperitoneally with 107 E. coli ATCC 25922, and subsequently euthanized at 24 h (if not already dead), after which the livers and spleens collected, homogenized and plated for c.f.u. analysis. Wild-type E. coli caused 60% death and showed high c.f.u. burdens in liver and spleen. All three darobactin-resistant bamA mutant strains had reduced virulence, with 100% survival in all groups at 24 h. The burden of bacteria of the triple bamA mutant was close to the limit of detection in organs, the G429R-expressing mutant was found at low but detectable levels, whereas the G429V-expressing mutant was found at relatively high loads in the organs. n = 5. Data are mean ± s.d. c, Schematic of the BAM activity assay in which BAM (BamA–E) was first inserted into lipid nanodiscs. Unfolded OmpT, along with the periplasmic chaperone SurA, was then mixed with the BAM–nanodiscs, and BAM folds OmpT into the nanodisc. OmpT, a protease, cleaves an internally quenched peptide, which produces a fluorescent signal. d, BAM–nanodisc assays performed in the presence of increasing concentrations of darobactin (left). The results show that darobactin is able to specifically inhibit BAM–nanodisc activity in a dose-dependent manner. These data were then normalized against the ‘no darobactin’ sample and the highest concentration of darobactin and plotted, and an IC50 was calculated using the online IC50 calculator tool (AAT Bioquest) (right). ND, nanodisc. n = 3 biologically independent experiments. Data are mean ± s.d. e, As a control to the BAM–nanodisc assays, we prepared OmpT–nanodiscs and assayed OmpT–nanodisc activity in the presence of increasing concentrations of darobactin. To prepare the OmpT–nanodiscs, we first expressed OmpT as inclusion bodies and then refolded the protein using previously reported methods55,56. We then incorporated OmpT into nanodiscs using the same methods as described for BAM. The assays were performed using 0.4 μM of OmpT–nanodiscs. The results show that darobactin has almost no effect on OmpT–nanodisc activity, thus confirming that darobactin does not affect OmpT activity itself or disrupting the nanodiscs themselves. A representative plot is shown from a triplicate experiment. f, The WNWSKSF peptide does not inhibit BAM–nanodiscs. As a control to darobactin, the BAM–nanodisc assays were performed in the presence of increasing concentrations of a linear peptide WNWSKSF. The results show that the WNWSKSF peptide has only minimal effects on BAM–nanodisc activity, even at the highest concentrations. A representative plot is shown from a triplicate experiment. g, h, Specific binding of darobactin to BamA/BAM. Mole ratio is the protein:ligand ratio. g, Plot of ITC experiments of wild-type BAM titrated with darobactin. Kd = 1.2 μM, N = 0.52, ΔH = –25 kcal mol−1 and ΔS = −56 cal mol−1 K−1. The experiment was repeated independently twice with similar results. h, Plot of ITC experiments of wild-type BAM titrated with the peptide WNWSKSF shows that there is no binding within the same concentration range as was used for darobactin. The experiment was repeated independently twice with similar results. i, j, Two-dimensional [15N, 1H]-TROSY spectra of 250 μM BamA-β in 0.1% w/v LDAO. i, BamA-β in the absence (left) and in the presence of darobactin with a molar ratio of 1:0.5 (middle) and 1:1 (right) of BamA-β:darobactin. The red dashed line outlines an example spectral region that shows substantial spectral changes during the titration. The experiment was repeated independently twice with similar results. j, An overlay of apo BamA-β (black) (250 μM) on BamA-β and a scrambled linear peptide WNKWSFS (green) (230 μM). The experiment was performed once.
Extended Data Fig. 6 Darobactin disrupts the outer membrane and causes lysis of E. coli.
E. coli MG1655 cells were placed on top of an agarose pad that contained darobactin and the fluorescent dyes FM4-64—to stain the membrane (false-coloured in magenta)—and Sytox Green—to show membrane permeabilization (false-coloured in green). E. coli MG1655 cells were observed over time at 37 °C under the microscope. For each indicated time point, representative panels show the killing progression of E. coli MG1655 with darobactin. White arrows highlight membrane blebbing; orange arrows highlight swelling and lysis. Scale bars, 5 μm. This figure is representative of three biologically independent experiments performed with similar results.
Extended Data Fig. 7 Transcriptome analysis of darobactin treatment shows activation of envelope stress pathways.
E. coli BW25113 were treated with 1× MIC darobactin, and the RNA isolated and sequenced. a–c, Volcano plots illustrate differential gene expression (Fisher’s exact test in edgeR; results were deemed significant if |log2(FC)| ≥ 2 and FDR-corrected P < 0.001; n = 3 biologically independent samples for each control or treatment sample) at time points t = 15 min (a), t = 30 min (b) and t = 60 min (c) after exposure. Grey region, not significant. d, Network visualization of differentially expressed genes at each time point. Nodes include genes (coloured circles) and time points (grey rectangles). Gene node colours represent relevant functional categories. Directed edges radiating from a time point node represent differentially expressed genes with respect to the given time point with weights reflecting the |log2(FC)|. e, Right, heat map showing the differential expression (|log2(FC)|) of genes of interest. Left, assignment to envelope stress pathways. Solid lines depict members of the same operon. In all panels, red indicates downregulation (lower expression in treatment relative to control) and blue indicates upregulation.
Extended Data Fig. 8 Two-dimensional thermal proteome profiling of darobactin.
a–c, Pseudo-volcano plots for two-dimensional thermal proteome profiling experiments of darobactin treatment (10 min) of E. coli BW25113 in living cells (a), lysates (b) and living cells pre-treated with chloramphenicol to inhibit protein synthesis (c). n = 1 for each concentration, heated to 10 different temperatures, for each experiment. Significant hits (FDR-adjusted P < 1%, calculated with a functional analysis of dose–response, requiring stabilization effects at n > 1 temperatures as described previously51) are highlighted in blue and integral outer membrane proteins are highlighted in purple. d, Heat maps for selected proteins in the experiment with living cells. For each protein and temperature (a key is shown on the right), the signal intensity was normalized to the vehicle control. e, Schematic of putative thermally stable assembled versus labile unassembled populations of the BAM machinery with darobactin treatment.
Extended Data Fig. 9 Darobactin single-dose pharmacokinetics and mouse thigh models.
a, Three mice were intraperitoneally injected with 50 mg kg−1 darobactin, and blood samples were collected by tail snip over 24 h. Samples (n = 1 per time point and mouse) were analysed for darobactin content by LC–MS/MS, and concentrations were calculated using a standard curve created by linear regression on the log(area under the curve peak) to log(concentration) of standards. Pharmacokinetic values were calculated in Excel; t1/2 and time > MIC assuming first-order elimination and using linear regression on time points 3–8 h; AUC (0–16 h) using the trapezoid rule. The limit of detection (LOD) was 0.08 μg ml−1. b, A mouse thigh model was repeated three times testing the efficacy of darobactin against E. coli AR350. Mice were injected with bacteria in their right thigh at 0 h, then dosed with no drug, gentamicin or darobactin starting 2 h after infection (50 mg kg−1 once, 25 mg kg−1 given three times every 6 h, or 20 mg kg−1 once). At 26 h mice were euthanized and thighs collected and homogenized tissues were plated for c.f.u. analysis. Data are mean ± s.d.
Supplementary information
Supplementary Discussion
Extra comments on screen strategy, NMR stereochemistry assignment, and on transcriptomics and proteomics results in Extended Data Figures 7 and 8.
41586_2019_1791_MOESM3_ESM.pdf
Supplementary Figure 1 Raw DNA gel for the dar operon knock out construction. Raw image of the agarose gel confirming the dar operon knockout in Extended Data Fig. 4c.
41586_2019_1791_MOESM4_ESM.zip
Supplementary Data 1 Raw NMR Data for BamA-darobactin titration and BamA-scramble peptide experiments. ZIP file containing raw NMR data related to Extended Data Figure 5i,j.
41586_2019_1791_MOESM5_ESM.pdf
Supplementary Table 1 Potency of darobactin against recent clinical isolates. Determination of darobactin and gentamicin MIC by broth microdilution was performed using broth microdilution by JMI laboratories (North Liberty, IA, USA).
41586_2019_1791_MOESM6_ESM.xlsx
Supplementary Table 2 Thermal Proteome Profiling Results. E. coli BW25113 living cells, with or without chloramphenicol pretreatment, or cell lysate, was treated with darobactin at five different concentrations. Aliquots of treated cells or lysates were then heated to ten different temperatures, and soluble proteins then analyzed by liquid chromatography coupled to tandem mass spectrometry (n=1 at each concentration for each experiment). Protein identification and quantification was performed using IsobarQuant and Mascot 2.4 (Matrix Science) against the E. coli Uniprot FASTA (Proteome ID: UP000000625). Data was analyzed with the TPP package for R followed by an FDR-controlled method for functional analysis of dose-response curves (see method reference). Supplementary Table 2 is related to Extended Data Figure 8.
41586_2019_1791_MOESM7_ESM.mov
Video 1 Time-lapse microscopy of darobactin causing outer membrane disruption and lysis of E. coli. E. coli MG1655 cells cultured in MHIIB were placed on top of a MHIIB/darobactin agarose pad containing FM4-64 and Sytox Green dyes, and observed under the microscope. Acquisition recording DIC, FM4-64 (false-coloured in magenta) and Sytox Green (false-coloured in green) signals was performed every 30 minutes at 37 °C for 14 hours. Scale bar, 10 μm. This figure is representative of three biologically independent experiments performed with similar results.
Rights and permissions
About this article
Cite this article
Imai, Y., Meyer, K.J., Iinishi, A. et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019). https://doi.org/10.1038/s41586-019-1791-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1791-1
This article is cited by
-
Tumescenamide C, a cyclic lipodepsipeptide from Streptomyces sp. KUSC_F05, exerts antimicrobial activity against the scab-forming actinomycete Streptomyces scabiei
The Journal of Antibiotics (2024)
-
A new type of antibiotic targets a drug-resistant bacterium
Nature (2024)
-
Engineered probiotic overcomes pathogen defences using signal interference and antibiotic production to treat infection in mice
Nature Microbiology (2024)
-
Non-modular fatty acid synthases yield distinct N-terminal acylation in ribosomal peptides
Nature Chemistry (2024)
-
Antibacterial coating on magnesium alloys by MAO for biomedical applications
Research on Biomedical Engineering (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.