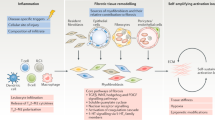
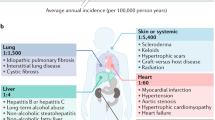
Abstract
Fibrosis can affect any organ and is responsible for up to 45% of all deaths in the industrialized world. It has long been thought to be relentlessly progressive and irreversible, but both preclinical models and clinical trials in various organ systems have shown that fibrosis is a highly dynamic process. This has clear implications for therapeutic interventions that are designed to capitalize on this inherent plasticity. However, despite substantial progress in our understanding of the pathobiology of fibrosis, a translational gap remains between the identification of putative antifibrotic targets and conversion of this knowledge into effective treatments in humans. Here we discuss the transformative experimental strategies that are being leveraged to dissect the key cellular and molecular mechanisms that regulate fibrosis, and the translational approaches that are enabling the emergence of precision medicine-based therapies for patients with fibrosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Eming, S. A., Martin, P. & Tomic-Canic, M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 6, 265sr6 (2014).
Allen, R. J. et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir. Med. 5, 869–880 (2017).
Kim, H. Y. et al. Genotype-related clinical characteristics and myocardial fibrosis and their association with prognosis in hypertrophic cardiomyopathy. J. Clin. Med. 9, E1671 (2020).
Young, C. N. J. et al. Total absence of dystrophin expression exacerbates ectopic myofiber calcification and fibrosis and alters macrophage infiltration patterns. Am. J. Pathol. 190, 190–205 (2020).
Schiller, H. B. et al. The Human Lung Cell Atlas: a high-resolution reference map of the human lung in health and disease. Am. J. Respir. Cell Mol. Biol. 61, 31–41 (2019).
Zepp, J. A. et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134–1148 (2017).
Xie, T. et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640 (2018).
Peyser, R. et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am. J. Respir. Cell Mol. Biol. 61, 74–85 (2019).
Tsukui, T. et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920 (2020).
Reyfman, P. A. et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199, 1517–1536 (2019).
Misharin, A. V. et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 214, 2387–2404 (2017).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Wu, H. et al. Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell 180, 107–121 (2020).
Adams, T. S. et al. Single cell RNA-seq reveals ectopic and aberrant lung resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2019).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Aizarani, N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Friedman, S. L., Roll, F. J., Boyles, J. & Bissell, D. M. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc. Natl Acad. Sci. USA 82, 8681–8685 (1985).
Dobie, R. et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 29, 1832–1847 (2019).
Krenkel, O., Hundertmark, J., Ritz, T. P., Weiskirchen, R. & Tacke, F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells 8, E503 (2019).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). This study dissected unanticipated aspects of the cellular and molecular basis of human liver fibrosis at a single-cell level, providing a framework for the discovery of rational therapeutic targets in liver cirrhosis.
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353 (2018).
Efremova, M. & Teichmann, S. A. Computational methods for single-cell omics across modalities. Nat. Methods 17, 14–17 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Ratziu, V. & Friedman, S. L. Why do so many NASH trials fail? Gastroenterology https://doi.org/10.1053/j.gastro.2020.05.046 (2020).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386 (2018).
Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019). This study uncovered anatomically discrete, functionally distinct subsets of fibroblasts in the context of arthritis.
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA 117, 15874–15883 (2020).
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature (in the press, 2020).
Der, E. et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 20, 915–927 (2019).
Driskell, R. R. et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281 (2013).
Rinkevich, Y. et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151 (2015).
Guerrero-Juarez, C. F. et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 10, 650 (2019).
Bergmeier, V. et al. Identification of a myofibroblast-specific expression signature in skin wounds. Matrix Biol. 65, 59–74 (2018).
Correa-Gallegos, D. et al. Patch repair of deep wounds by mobilized fascia. Nature 576, 287–292 (2019). This work identified a specialized subset of fibroblasts, fascia fibroblasts, which gather the surrounding ECM and then rise to the surface of the skin after wounding.
Shook, B. A. et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971 (2018).
Montero-Melendez, T. et al. Therapeutic senescence via GPCR activation in synovial fibroblasts facilitates resolution of arthritis. Nat. Commun. 11, 745 (2020).
Schafer, M. J., Haak, A. J., Tschumperlin, D. J. & LeBrasseur, N. K. Targeting senescent cells in fibrosis: pathology, paradox, and practical considerations. Curr. Rheumatol. Rep. 20, 3 (2018).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020).
Hickson, L. J. et al. Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine 47, 446–456 (2019).
Schneider, R. K. et al. Gli1+ mesenchymal stromal cells are a key driver of bone marrow fibrosis and an important cellular therapeutic target. Cell Stem Cell 23, 308–309 (2018).
El Agha, E. et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20, 261–273 (2017).
Scott, R. W., Arostegui, M., Schweitzer, R., Rossi, F. M. V. & Underhill, T. M. Hic1 defines quiescent mesenchymal progenitor subpopulations with distinct functions and fates in skeletal muscle regeneration. Cell Stem Cell 25, 797–813 (2019).
Soliman, H. et al. Pathogenic potential of Hic1-expressing cardiac stromal progenitors. Cell Stem Cell 26, 459–461 (2020).
Mahmoudi, S. et al. Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature 574, 553–558 (2019).
Kisseleva, T. et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl Acad. Sci. USA 109, 9448–9453 (2012).
Troeger, J. S. et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143, 1073–1083 (2012).
Wohlfahrt, T. et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature 566, 344–349 (2019).
Plikus, M. V. et al. Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752 (2017).
Song, G. et al. Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell 18, 797–808 (2016).
Rezvani, M. et al. In vivo hepatic reprogramming of myofibroblasts with AAV vectors as a therapeutic strategy for liver fibrosis. Cell Stem Cell 18, 809–816 (2016).
Aghajanian, H. et al. Targeting cardiac fibrosis with engineered T cells. Nature 573, 430–433 (2019).
Pereira, B. I. et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 10, 2387 (2019).
Pakshir, P. & Hinz, B. The big five in fibrosis: macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 68-69, 81–93 (2018).
Januszyk, M. et al. Mechanical offloading of incisional wounds is associated with transcriptional downregulation of inflammatory pathways in a large animal model. Organogenesis 10, 186–193 (2014).
Froese, A. R. et al. Stretch-induced activation of transforming growth factor-β1 in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 194, 84–96 (2016).
Lindsey, M. L., Iyer, R. P., Jung, M., DeLeon-Pennell, K. Y. & Ma, Y. Matrix metalloproteinases as input and output signals for post-myocardial infarction remodeling. J. Mol. Cell. Cardiol. 91, 134–140 (2016).
Craig, V. J., Zhang, L., Hagood, J. S. & Owen, C. A. Matrix metalloproteinases as therapeutic targets for idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 53, 585–600 (2015).
Lundberg, E. & Borner, G. H. H. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 20, 285–302 (2019).
Schwabe, R. F., Tabas, I. & Pajvani, U. B. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology 158, 1913–1928 (2020).
Xie, N. et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am. J. Respir. Crit. Care Med. 192, 1462–1474 (2015).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371 (2017).
Kottmann, R. M. et al. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-β. Am. J. Respir. Crit. Care Med. 186, 740–751 (2012).
Liu, G. & Summer, R. Cellular metabolism in lung health and disease. Annu. Rev. Physiol. 81, 403–428 (2019).
Nigdelioglu, R. et al. Transforming growth factor (TGF)-β promotes de novo serine synthesis for collagen production. J. Biol. Chem. 291, 27239–27251 (2016).
Park, S. Y., Le, C. T., Sung, K. Y., Choi, D. H. & Cho, E. H. Succinate induces hepatic fibrogenesis by promoting activation, proliferation, and migration, and inhibiting apoptosis of hepatic stellate cells. Biochem. Biophys. Res. Commun. 496, 673–678 (2018).
Lian, N. et al. Curcumin regulates cell fate and metabolism by inhibiting hedgehog signaling in hepatic stellate cells. Lab. Invest. 95, 790–803 (2015).
Ding, H. et al. Inhibiting aerobic glycolysis suppresses renal interstitial fibroblast activation and renal fibrosis. Am. J. Physiol. Renal Physiol. 313, F561–F575 (2017).
Wei, Q. et al. Glycolysis inhibitors suppress renal interstitial fibrosis via divergent effects on fibroblasts and tubular cells. Am. J. Physiol. Renal Physiol. 316, F1162–F1172 (2019).
Ge, J. et al. Glutaminolysis promotes collagen translation and stability via α-ketoglutarate-mediated mTOR activation and proline hydroxylation. Am. J. Respir. Cell Mol. Biol. 58, 378–390 (2018).
Bai, L. et al. Glutaminolysis epigenetically regulates antiapoptotic gene expression in idiopathic pulmonary fibrosis fibroblasts. Am. J. Respir. Cell Mol. Biol. 60, 49–57 (2019).
Cui, H. et al. Inhibition of glutaminase 1 attenuates experimental pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 61, 492–500 (2019).
Kang, H. M. et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21, 37–46 (2015). This study elegantly links abnormal fatty acid oxidation to fibrogenesis.
Luengo, A., Gui, D. Y. & Vander Heiden, M. G. Targeting metabolism for cancer therapy. Cell Chem. Biol. 24, 1161–1180 (2017).
Wynn, T. A. & Vannella, K. M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462 (2016).
Vannella, K. M. & Wynn, T. A. Mechanisms of organ injury and repair by macrophages. Annu. Rev. Physiol. 79, 593–617 (2017).
Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017).
Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019).
Guilliams, M., Thierry, G. R., Bonnardel, J. & Bajenoff, M. Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020).
Lavine, K. J. et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl Acad. Sci. USA 111, 16029–16034 (2014).
Duffield, J. S. et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Invest. 115, 56–65 (2005).
Borthwick, L. A. et al. Macrophages are critical to the maintenance of IL-13-dependent lung inflammation and fibrosis. Mucosal Immunol. 9, 38–55 (2016).
Satoh, T. et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017).
Bajpai, G. et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 24, 1234–1245 (2018).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Dick, S. A. et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39 (2019).
Wang, J. & Kubes, P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 165, 668–678 (2016).
Deniset, J. F. et al. Gata6+ pericardial cavity macrophages relocate to the injured heart and prevent cardiac fibrosis. Immunity 51, 131–140 (2019).
Adler, M. et al. Principles of cell circuits for tissue repair and fibrosis. iScience 23, 100841 (2020).
Henderson, N. C. et al. Galectin-3 expression and secretion links macrophages to the promotion of renal fibrosis. Am. J. Pathol. 172, 288–298 (2008).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757 (2018).
Lodyga, M. et al. Cadherin-11-mediated adhesion of macrophages to myofibroblasts establishes a profibrotic niche of active TGFβ. Sci. Signal. 12, eaao3469 (2019).
Minutti, C. M. et al. A macrophage-pericyte axis directs tissue restoration via amphiregulin-induced transforming growth factor beta activation. Immunity 50, 645–654 (2019).
Pakshir, P. et al. Dynamic fibroblast contractions attract remote macrophages in fibrillar collagen matrix. Nat. Commun. 10, 1850 (2019).
Diebold, R. J. et al. Early-onset multifocal inflammation in the transforming growth factor beta 1-null mouse is lymphocyte mediated. Proc. Natl Acad. Sci. USA 92, 12215–12219 (1995).
McEntee, C. P., Gunaltay, S. & Travis, M. A. Regulation of barrier immunity and homeostasis by integrin-mediated transforming growth factor β activation. Immunology 160, 139–148 (2020).
Kelly, A. et al. Human monocytes and macrophages regulate immune tolerance via integrin αvβ8-mediated TGFβ activation. J. Exp. Med. 215, 2725–2736 (2018).
Barczyk, M., Carracedo, S. & Gullberg, D. Integrins. Cell Tissue Res. 339, 269–280 (2010).
Hynes, R. O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002).
Robertson, I. B. & Rifkin, D. B. Regulation of the bioavailability of TGFβ and TGFβ-related proteins. Cold Spring Harb. Perspect. Biol. 8, a021907 (2016).
Reed, N. I. et al. The αvβ1 integrin plays a critical in vivo role in tissue fibrosis. Sci. Transl. Med. 7, 288ra79 (2015).
Munger, J. S. et al. A mechanism for regulating pulmonary inflammation and fibrosis: the integrin αvβ6 binds and activates latent TGFβ1. Cell 96, 319–328 (1999).
Wipff, P. J., Rifkin, D. B., Meister, J. J. & Hinz, B. Myofibroblast contraction activates latent TGFβ1 from the extracellular matrix. J. Cell Biol. 179, 1311–1323 (2007).
Shi, M. et al. Latent TGFβ structure and activation. Nature 474, 343–349 (2011).
Dong, X. et al. Force interacts with macromolecular structure in activation of TGFβ. Nature 542, 55–59 (2017).
Dong, X., Hudson, N. E., Lu, C. & Springer, T. A. Structural determinants of integrin β-subunit specificity for latent TGFβ. Nat. Struct. Mol. Biol. 21, 1091–1096 (2014).
Campbell, M. G. et al. Cryo-EM reveals integrin-mediated TGFβ activation without release from latent TGFβ. Cell 180, 490–501 (2020).
Hahm, K. et al. αvβ6 integrin regulates renal fibrosis and inflammation in Alport mouse. Am. J. Pathol. 170, 110–125 (2007).
Wang, B. et al. Role of αvβ6 integrin in acute biliary fibrosis. Hepatology 46, 1404–1412 (2007).
Peng, Z. W. et al. Integrin αvβ6 critically regulates hepatic progenitor cell function and promotes ductular reaction, fibrosis, and tumorigenesis. Hepatology 63, 217–232 (2016).
Horan, G. S. et al. Partial inhibition of integrin αvβ6 prevents pulmonary fibrosis without exacerbating inflammation. Am. J. Respir. Crit. Care Med. 177, 56–65 (2008).
Puthawala, K. et al. Inhibition of integrin αvβ6, an activator of latent transforming growth factor-β, prevents radiation-induced lung fibrosis. Am. J. Respir. Crit. Care Med. 177, 82–90 (2008).
Araya, J. et al. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J. Clin. Invest. 117, 3551–3562 (2007).
Kitamura, H. et al. Mouse and human lung fibroblasts regulate dendritic cell trafficking, airway inflammation, and fibrosis through integrin αvβ8-mediated activation of TGFβ. J. Clin. Invest. 121, 2863–2875 (2011).
Minagawa, S. et al. Selective targeting of TGFβ activation to treat fibroinflammatory airway disease. Sci. Transl. Med. 6, 241ra79 (2014).
Henderson, N. C. et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 (2013).
Barron, L. & Wynn, T. A. Fibrosis is regulated by Th2 and Th17 responses and by dynamic interactions between fibroblasts and macrophages. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G723–G728 (2011).
Park, M. J. et al. IL-1–IL-17 signaling axis contributes to fibrosis and inflammation in two different murine models of systemic sclerosis. Front. Immunol. 9, 1611 (2018).
Wilson, M. S. et al. Bleomycin and IL-1β-mediated pulmonary fibrosis is IL-17A dependent. J. Exp. Med. 207, 535–552 (2010).
Wang, B. Z. et al. Interleukin-17A antagonist attenuates radiation-induced lung injuries in mice. Exp. Lung Res. 40, 77–85 (2014).
Meng, F. et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 143, 765–776 (2012).
Sun, B. et al. Role of interleukin 17 in TGFβ signaling-mediated renal interstitial fibrosis. Cytokine 106, 80–88 (2018).
Feng, W. et al. IL-17 induces myocardial fibrosis and enhances RANKL/OPG and MMP/TIMP signaling in isoproterenol-induced heart failure. Exp. Mol. Pathol. 87, 212–218 (2009).
Fabre, T. et al. Type 3 cytokines IL-17A and IL-22 drive TGFβ-dependent liver fibrosis. Sci. Immunol. 3, eaar7754 (2018).
Tan, Z. et al. IL-17A plays a critical role in the pathogenesis of liver fibrosis through hepatic stellate cell activation. J. Immunol. 191, 1835–1844 (2013).
Zhang, S. et al. Neutralization of interleukin-17 attenuates cholestatic liver fibrosis in mice. Scand. J. Immunol. 83, 102–108 (2016).
Zhang, X. W. et al. Antagonism of interleukin-17A ameliorates experimental hepatic fibrosis by restoring the IL-10/STAT3-suppressed autophagy in hepatocytes. Oncotarget 8, 9922–9934 (2017).
Fabre, T., Kared, H., Friedman, S. L. & Shoukry, N. H. IL-17A enhances the expression of profibrotic genes through upregulation of the TGFβ receptor on hepatic stellate cells in a JNK-dependent manner. J. Immunol. 193, 3925–3933 (2014).
Oh, K. et al. Epithelial transglutaminase 2 is needed for T cell interleukin-17 production and subsequent pulmonary inflammation and fibrosis in bleomycin-treated mice. J. Exp. Med. 208, 1707–1719 (2011).
Wree, A. et al. NLRP3 inflammasome driven liver injury and fibrosis: roles of IL-17 and TNF in mice. Hepatology 67, 736–749 (2018).
Gasse, P. et al. IL-1 and IL-23 mediate early IL-17A production in pulmonary inflammation leading to late fibrosis. PLoS One 6, e23185 (2011).
Lemmers, A. et al. The interleukin-17 pathway is involved in human alcoholic liver disease. Hepatology 49, 646–657 (2009).
Macek Jilkova, Z. et al. Progression of fibrosis in patients with chronic viral hepatitis is associated with IL-17+ neutrophils. Liver Int. 36, 1116–1124 (2016).
Yang, D. et al. Dysregulated lung commensal bacteria drive interleukin-17b production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity 50, 692–706 (2019).
Seki, E. et al. TLR4 enhances TGFβ signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 (2007).
de Kretser, D. M. et al. Serum activin A and B levels predict outcome in patients with acute respiratory failure: a prospective cohort study. Crit. Care 17, R263 (2013).
Gieseck, R. L., III, Wilson, M. S. & Wynn, T. A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18, 62–76 (2018).
Hams, E. et al. IL-25 and type 2 innate lymphoid cells induce pulmonary fibrosis. Proc. Natl Acad. Sci. USA 111, 367–372 (2014).
Jessup, H. K. et al. Intradermal administration of thymic stromal lymphopoietin induces a T cell- and eosinophil-dependent systemic Th2 inflammatory response. J. Immunol. 181, 4311–4319 (2008).
McHedlidze, T. et al. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 39, 357–371 (2013).
Vannella, K. M. et al. Combinatorial targeting of TSLP, IL-25, and IL-33 in type 2 cytokine-driven inflammation and fibrosis. Sci. Transl. Med. 8, 337ra65 (2016).
Lee, C. G. et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor β1. J. Exp. Med. 194, 809–821 (2001).
Kaviratne, M. et al. IL-13 activates a mechanism of tissue fibrosis that is completely TGFβ independent. J. Immunol. 173, 4020–4029 (2004).
Gieseck, R. L. III et al. Interleukin-13 activates distinct cellular pathways leading to ductular reaction, steatosis, and fibrosis. Immunity 45, 145–158 (2016).
Hart, K. M. et al. Type 2 immunity is protective in metabolic disease but exacerbates NAFLD collaboratively with TGFβ. Sci. Transl. Med. 9, eaal3694 (2017). This study identified opposing roles for type 2 immunity in metabolic syndrome and liver fibrosis in an experimental model of NASH.
Chiaramonte, M. G., Donaldson, D. D., Cheever, A. W. & Wynn, T. A. An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J. Clin. Invest. 104, 777–785 (1999).
Xue, J. et al. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun. 6, 7158 (2015).
Liu, L. et al. CD4+ T lymphocytes, especially Th2 cells, contribute to the progress of renal fibrosis. Am. J. Nephrol. 36, 386–396 (2012).
Chung, S. I. et al. IL-13 is a therapeutic target in radiation lung injury. Sci. Rep. 6, 39714 (2016).
Singh, B., Kasam, R. K., Sontake, V., Wynn, T. A. & Madala, S. K. Repetitive intradermal bleomycin injections evoke T-helper cell 2 cytokine-driven pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L796–L806 (2017).
Sciurba, J. C. et al. Fibroblast-specific integrin-alpha V differentially regulates type 17 and type 2 driven inflammation and fibrosis. J. Pathol. 248, 16–29 (2019).
Wang, M. et al. Cross-talk between TH2 and TH17 pathways in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 144, 1254–1264 (2019).
Choy, D. F. et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci. Transl. Med. 7, 301ra129 (2015).
Ramalingam, T. R. et al. Enhanced protection from fibrosis and inflammation in the combined absence of IL-13 and IFN-γ. J. Pathol. 239, 344–354 (2016).
Tang, W. et al. Targeted expression of IL-11 in the murine airway causes lymphocytic inflammation, bronchial remodeling, and airways obstruction. J. Clin. Invest. 98, 2845–2853 (1996). This paper identified autocrine IL-11–IL-11R signaling in fibroblasts as a key mechanism driving cardiovascular fibrosis in response to a variety of stimuli.
Schafer, S. et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017).
Ng, B. et al. Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci. Transl. Med. 11, eaaw1237 (2019).
Abreu, M. T. et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn’s disease. Gastroenterology 123, 679–688 (2002).
Rieder, F. et al. Association of the novel serologic anti-glycan antibodies anti-laminarin and anti-chitin with complicated Crohn’s disease behavior. Inflamm. Bowel Dis. 16, 263–274 (2010).
Rieder, F. et al. Serum anti-glycan antibodies predict complicated Crohn’s disease behavior: a cohort study. Inflamm. Bowel Dis. 16, 1367–1375 (2010).
Rieder, F., Kessler, S., Sans, M. & Fiocchi, C. Animal models of intestinal fibrosis: new tools for the understanding of pathogenesis and therapy of human disease. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G786–G801 (2012).
Moresco, E. M., LaVine, D. & Beutler, B. Toll-like receptors. Curr. Biol. 21, R488–R493 (2011).
Månsson, L. E. et al. MyD88 signaling promotes both mucosal homeostatic and fibrotic responses during Salmonella-induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G311–G323 (2012).
Imai, J. et al. Flagellin-mediated activation of IL-33-ST2 signaling by a pathobiont promotes intestinal fibrosis. Mucosal Immunol. 12, 632–643 (2019).
Jacob, N. et al. Inflammation-independent TL1A-mediated intestinal fibrosis is dependent on the gut microbiome. Mucosal Immunol. 11, 1466–1476 (2018).
Otte, J. M., Rosenberg, I. M. & Podolsky, D. K. Intestinal myofibroblasts in innate immune responses of the intestine. Gastroenterology 124, 1866–1878 (2003).
Zhao, S. et al. Selective deletion of MyD88 signaling in α-SMA positive cells ameliorates experimental intestinal fibrosis via post-transcriptional regulation. Mucosal Immunol. 13, 665–678 (2020). This study highlights a selective mechanism by which bacteria activate myofibroblasts through flagellin.
Chan, C. C. et al. Prognostic value of plasma endotoxin levels in patients with cirrhosis. Scand. J. Gastroenterol. 32, 942–946 (1997).
Seki, E. & Brenner, D. A. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology 48, 322–335 (2008).
Sun, L. et al. Lipopolysaccharide enhances TGFβ1 signalling pathway and rat pancreatic fibrosis. J. Cell. Mol. Med. 22, 2346–2356 (2018).
Yang, L. et al. TRIF differentially regulates hepatic steatosis and inflammation/fibrosis in mice. Cell. Mol. Gastroenterol. Hepatol. 3, 469–483 (2017).
Mazagova, M. et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 29, 1043–1055 (2015).
Leaf, I. A. et al. Pericyte MyD88 and IRAK4 control inflammatory and fibrotic responses to tissue injury. J. Clin. Invest. 127, 321–334 (2017).
Jialal, I., Major, A. M. & Devaraj, S. Global Toll-like receptor 4 knockout results in decreased renal inflammation, fibrosis and podocytopathy. J. Diabetes Complications 28, 755–761 (2014).
Liu, J. H. et al. A novel inhibitor of homodimerization targeting MyD88 ameliorates renal interstitial fibrosis by counteracting TGFβ1-induced EMT in vivo and in vitro. Kidney Blood Press. Res. 43, 1677–1687 (2018).
Stifano, G. et al. Chronic Toll-like receptor 4 stimulation in skin induces inflammation, macrophage activation, transforming growth factor beta signature gene expression, and fibrosis. Arthritis Res. Ther. 16, R136 (2014).
Liang, J. et al. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat. Med. 22, 1285–1293 (2016). This work discovered an anti-fibrotic mechanism for hyaluronan in pulmonary fibrosis, revealing a novel function for TLR4.
Pilling, D. et al. Reduction of bleomycin-induced pulmonary fibrosis by serum amyloid P. J. Immunol. 179, 4035–4044 (2007).
Nakagawa, N. et al. Pentraxin-2 suppresses c-Jun/AP-1 signaling to inhibit progressive fibrotic disease. JCI Insight 1, e87446 (2016).
Rogliani, P., Calzetta, L., Cavalli, F., Matera, M. G. & Cazzola, M. Pirfenidone, nintedanib and N-acetylcysteine for the treatment of idiopathic pulmonary fibrosis: a systematic review and meta-analysis. Pulm. Pharmacol. Ther. 40, 95–103 (2016).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Eng, C. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Dudley, J. T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3, 96ra76 (2011).
Torok, N. J., Dranoff, J. A., Schuppan, D. & Friedman, S. L. Strategies and endpoints of antifibrotic drug trials: summary and recommendations from the AASLD Emerging Trends Conference, Chicago, June 2014. Hepatology 62, 627–634 (2015).
Rieder, F. et al. An expert consensus to standardise definitions, diagnosis and treatment targets for anti-fibrotic stricture therapies in Crohn’s disease. Aliment. Pharmacol. Ther. 48, 347–357 (2018).
Montesi, S. B., Désogère, P., Fuchs, B. C. & Caravan, P. Molecular imaging of fibrosis: recent advances and future directions. J. Clin. Invest. 129, 24–33 (2019).
Montesi, S. B. et al. Type I collagen-targeted positron emission tomography imaging in idiopathic pulmonary fibrosis: first-in-human studies. Am. J. Respir. Crit. Care Med. 200, 258–261 (2019).
Acknowledgements
N.C.H. is supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 219542/Z/19/Z), the Medical Research Council, a Chan Zuckerberg Initiative Seed Network Grant, the British Heart Foundation and Tenovus Scotland. F.R. is supported by grants from the National Institutes of Health (T32DK083251, P30DK097948 Pilot, K08DK110415 and R01DK123233), the Crohn’s and Colitis Foundation, the Cleveland Clinic, the Rainin Foundation and the Helmsley Charitable Trust through the Stenosis Therapy and Anti-Fibrotic Research (STAR) Consortium.
Author information
Authors and Affiliations
Contributions
N.C.H., F.R. and T.A.W contributed equally to the writing and editing of all aspects of this review.
Corresponding author
Ethics declarations
Competing interests
N.C.H. has received research funding from AbbVie, Pfizer, Gilead and Galecto, and is an advisor or consultant for Galecto, Indalo Therapeutics, Pliant Therapeutics, GSK and Boehringer-Ingelheim. F.R. is an advisor or consultant for AbbVie, Allergan, BMS, Boehringer-Ingelheim, Celgene, Falk Pharma, Gilead, Genentech, Gossamer, GSK, Receptos, Thetis, UCB, Samsung, Koutif, Pliant Therapeutics, Metacrine, Takeda, Theravance, Pfizer, Agomab, Helmsley, RedX and Roche. T.A.W. is employed by Pfizer.
Additional information
Peer review information Nature thanks Christopher Buckley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2.
Rights and permissions
About this article
Cite this article
Henderson, N.C., Rieder, F. & Wynn, T.A. Fibrosis: from mechanisms to medicines. Nature 587, 555–566 (2020). https://doi.org/10.1038/s41586-020-2938-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2938-9
This article is cited by
-
The occurrence and development mechanisms of esophageal stricture: state of the art review
Journal of Translational Medicine (2024)
-
HWJMSC-EVs promote cartilage regeneration and repair via the ITGB1/TGF-β/Smad2/3 axis mediated by microfractures
Journal of Nanobiotechnology (2024)
-
Inhibitor of PD-1/PD-L1: a new approach may be beneficial for the treatment of idiopathic pulmonary fibrosis
Journal of Translational Medicine (2024)
-
Fibroblasts inhibit osteogenesis by regulating nuclear-cytoplasmic shuttling of YAP in mesenchymal stem cells and secreting DKK1
Biological Research (2024)
-
Fibroblast and myofibroblast activation in normal tissue repair and fibrosis
Nature Reviews Molecular Cell Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.