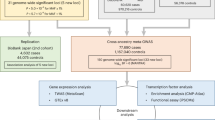
Abstract
Atrial fibrillation (AF) affects more than 33 million individuals worldwide1 and has a complex heritability2. We conducted the largest meta-analysis of genome-wide association studies (GWAS) for AF to date, consisting of more than half a million individuals, including 65,446 with AF. In total, we identified 97 loci significantly associated with AF, including 67 that were novel in a combined-ancestry analysis, and 3 that were novel in a European-specific analysis. We sought to identify AF-associated genes at the GWAS loci by performing RNA-sequencing and expression quantitative trait locus analyses in 101 left atrial samples, the most relevant tissue for AF. We also performed transcriptome-wide analyses that identified 57 AF-associated genes, 42 of which overlap with GWAS loci. The identified loci implicate genes enriched within cardiac developmental, electrophysiological, contractile and structural pathways. These results extend our understanding of the biological pathways underlying AF and may facilitate the development of therapeutics for AF.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
20 July 2018
In the version of this article initially published, Supplementary Tables 1, 2, 6, 8, 10 and 19–22 and the Supplementary Note were omitted from the supplementary PDF. The supplementary PDF now includes these items.
References
Chugh, S. S. et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 study. Circulation 129, 837–847 (2014).
Lubitz, S. A. et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 304, 2263–2269 (2010).
January, C. T. et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: executive summary. J. Am. Coll. Cardiol. 64, 2071–2104 (2014).
Benjamin, E. J. et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat. Genet. 41, 879–881 (2009).
Ellinor, P. T. et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 44, 670–675 (2012).
Sinner, M. F. et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 130, 1225–1235 (2014).
Ellinor, P. T. et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet. 42, 240–244 (2010).
Christophersen, I. E. et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 49, 946–952 (2017).
Low, S.-K. et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat. Genet. 49, 953–958 (2017).
Weng, L.-C. et al. Heritability of atrial fibrillation. Circ. Cardiovasc. Genet. 10, e001838 (2017).
Klarin, D. et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet. 49, 1392–1397 (2017).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Lu, X. et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum. Mol. Genet. 24, 865–74 (2015).
Nielsen, J. B. et al. Genome-wide association study of 1 million people identifies 111 loci for atrial fibrillation. Preprint at https://www.biorxiv.org/content/early/2018/01/04/242149 (2018).
Sinner, M. F. et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG). Eur. Heart J. 29, 907–914 (2008).
Olson, T. M. et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 293, 447–454 (2005).
McNair, W. P. et al. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation 110, 2163–2167 (2004).
van Weerd, J. H. et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ. Res. 115, 432–441 (2014).
Schott, J. J. et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281, 108–101 (1998).
den Hoed, M. et al. Identification of heart rate–associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631 (2013).
Kirchhof, P. et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ. Cardiovasc. Genet. 4, 123–133 (2011).
Wang, J. et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA 107, 9753–9758 (2010).
Syeda, F. et al. PITX2 modulates atrial membrane potential and the antiarrhythmic effects of sodium-channel blockers. J. Am. Coll. Cardiol. 68, 1881–1894 (2016).
Nadadur, R. D. et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci. Transl. Med. 8, 354ra115 (2016).
Tucker, N. R. et al. Diminished PRRX1 expression is associated with increased risk of atrial fibrillation and shortening of the cardiac action potential. Circ. Cardiovasc. Genet. 10, e001902 (2017).
Postma, A. V. et al. A gain-of-function TBX5 mutation is associated with atypical Holt–Oram syndrome and paroxysmal atrial fibrillation. Circ. Res. 102, 1433–1442 (2008).
Lahat, H. et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am. J. Hum. Genet. 69, 1378–1384 (2001).
Lahat, H. et al. Autosomal recessive catecholamine- or exercise-induced polymorphic ventricular tachycardia: clinical features and assignment of the disease gene to chromosome 1p13-21. Circulation 103, 2822–2827 (2001).
Corrado, D., Link, M. S. & Calkins, H. Arrhythmogenic right ventricular cardiomyopathy. N. Engl. J. Med. 376, 61–72 (2017).
Gerull, B. et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 36, 1162–1164 (2004).
Ackerman, M. J. et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies. Heart Rhythm 8, 1308–1339 (2011).
Weng, L.-C. et al. Genetic predisposition, clinical risk factor burden, and lifetime risk of atrial fibrillation. Circulation 137, 1027–1038 (2017).
Korn, J. M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008).
Goldstein, J. I. et al. zCall: a rare variant caller for array-based genotyping. Bioinformatics 28, 2543–2545 (2012).
Pulit, S. L. et al. Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 15, 174–184 (2016).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at https://www.biorxiv.org/content/early/2017/07/20/166298 (2017).
The Haplotype Reference Consortium et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Francioli, L. C. et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Bellenguez, C., Strange, A., Freeman, C., Donnelly, P. & Spencer, C. C. A. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012).
Aulchenko, Y. S., Struchalin, M. V. & van Duijn, C. M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Chanda, P., Huang, H., Arking, D. E. & Bader, J. S. Fast association tests for genes with FAST. PLoS One 8, e68585 (2013).
R Core Team. R: A Language and Environment for Statistical Computing. http://www.r-project.org/ (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Fadista, J., Manning, A. K., Florez, J. C. & Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016).
Higgins, J. P. T., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. Br. Med. J. 327, 557–560 (2003).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ernst, J. & Kellis, M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 33, 364–376 (2015).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Pers, T. H., Timshel, P. & Hirschhorn, J. N. SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics 31, 418–420 (2015).
Fay, M. P. & Shaw, P. A. Exact and asymptotic weighted logrank tests for interval censored data: the interval R package. J. Stat. Softw. 36, 1–34 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Harrow, J. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Delaneau, O. et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017).
GTEx Consortium. The Genotype–Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Segrè, A. V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Loh, P.-R. et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 47, 1385–1392 (2015).
Acknowledgements
A full list of acknowledgments appears in the Supplementary Notes.
Author information
Authors and Affiliations
Contributions
C.R., M.D.C., E.J.B., K.L.L., S.A.L., P.T.E. and H.L. drafted and finalized the manuscript. H.J.C., E.A.D., B.L.K., B. Weijs, S. Kääb, M.M.-N., B.N., K.S., M.F.S., J.L., A.A., L.Y.C., K.L., S.A., D.C., G.P., L. Risch, S. Thériault, T.T., C. Schurman, S.A.S., J.C.D., D.M.R., Q.S.W., C.R., M.D.C., K.G.A., B.R.D., N.G., S. Kathiresan, L.M., P.L.H., J.B., M.K.C., J.D.S., H. Sun, D.R.V.W., T.M.B., J.C.B., J.A.B., G.A., M.S.O., L. Refsgaard, J.H.S., D.F., R.J., S. Shah, P.K., R.B.S., T.E., M.T.-L., E.J.B., B. Wang, K.L.L., M. Kähönen, T.L., I.E.C., I.C.V.G., B.G., M. Rienstra, J.E.S., P.V.D.H., N.V., H.L.B., S.C.D., R.G., B.L., S. Saba, A.A.S., R.W., H.C., R.N.L., N.L.S., K.L.W., S.R.H., B.M.P., N.S., J. Carlquist, M.J.C., S. Knight, M.E.K., W.M., P.A., O.M., M.O.-M., X.G., H.J.L., J.I.R., K.D.T., S.H.C., N.R.T., S.A.L., P.T.E., C.N.-C., M.A.R., C.D.A., P.N., J.J.G.S., H. Schunkert, T.P.C., K.B.M., I.F., J.J.W.J., P.W.M., R.N., S. Trompet, O.H.F., A. Hofman, M. Kavousi, M.N.N., B.H. Stricker, A.G.U., R.P.G., J.J.-C., S.L.P., S.M., A. Hamsten, J.P.K., G.M.M., C.R.P., A.P.M., S.G., E. Ingelsson, H.L., D.D., J.A.M., M.M.B.S., Z.T.Y., C. Shaffer, P.E.W., C.M.A., D.I.C., R.K.S., J.W., M. Dichgans and R.M. contributed to and revised the manuscript. H.J.C., E.A.D., B.L.K., B. Weijs, S. Kääb, M.M.-N., B.N., K.S., M.F.S., V.G., T.B.H., L.J.L., A.V.S., M.E., J. Hernesniemi, J.L., I.S., A.A., D.E.A., N.A.B., E.B., L.Y.C., M.L., E.Z.S., S.A., D.C., G.P., L. Risch, S. Thériault, K.I., Y.K., M. Kubo, S.-K.L., T.T., E.B.B., R.J.F.L., Y.L., C. Schurman, S.A.S., J.C.D., D.M.R., Q.S.W., C.R., M.D.C., L.-C.W., K.G.A., N.G., S. Kathiresan, L.M., P.L.H., J.B., M.K.C., J.D.S., H. Sun, D.R.V.W., T.M.B., J.C.B., J.A.B., M.-L.L., J. Sinisalo, E.V., G.A., M.S.O., L. Refsgaard, J.H.S., D.F., R.J., A. Sun, P.K., H.O., R.B.S., T.Z., T.E., M.T.-L., E.J.B., B. Wang, K.L.L., M. Kähönen, T.L., L.-P.L., K.N., I.E.C., A. Tveit, B.G., J.E.S., N.V., H.L.B., S.C.D., R.G., B.L., S. Saba, A.A.S., R.W., A.C., C.H., L.J.H., J. Huffman, S.P., D.P., B.H. Smith, H.C., E. Ipek, S.N., R.N.L., N.L.S., K.L.W., S.R.H., B.M.P., N.S., J. Carlquist, M.J.C., S. Knight, E.-K.C., H.E.L., H.-N.P., J. Shim, P.-S.Y., G.D., J. Huang, M.E.K., P.A., O.M., M.O.-M., Y.-D.C., X.G., K.D.T., J.Y., S.A.L., P.T.E., C.N.-C., M.A.R., J.R., N.R., C.D.A., P.N., J.J.G.S., A.K., T.K., H. Schunkert, L.Z., T.P.C., S.M.D., K.B.M., M.P.M., D.J.R., I.F., J.J.W.J., S. Trompet, O.H.F., A. Hofman, M. Kavousi, M.N.N., B.H. Stricker, A.G.U., M. Dörr, S.B.F., A. Teumer, U.V., S.W., J.W.C., R.P.G., J.J.-C., P.K.-W., J.P., S.L.P., M. Ribasés, A. Slowik, D.W., B.B.W., A.R.V.R.H., J.E.K., A.J.M., A.P., S.M., A.N., A. Hamsten, P.K.M., N.L.P., J.P.K., G.M.M., C.R.P., J. Cook, L.L., C.M.L., A.M., A.P.M., S.G., E. Ingelsson, N.E., K.T., H.L., D.D.M., D.D., J.A.M., M.M.B.S., Z.T.Y., C. Shaffer, P.E.W., C.M.A., D.I.C., P.M.R., M. Dichgans and R.M. contributed to study-specific GWAS by providing phenotype data or performing data analyses. C.R., M.D.C. and S.L.P. performed meta-analyses. N.R.T., P.T.E., T.P.C., K.B.M., M.P.M. and H.L. contributed samples sequencing or performed left atrial eQTL analyses. C.R., M.D.C., L.-C.W., K.L.L., S.H.C., N.R.T. and H.L. performed downstream analyses. K.I., T.T., K.L.L., S.R.H., S.A.L. and P.T.E. conceived designed and supervised the overall project.
Corresponding author
Ethics declarations
Competing interests
P.T.E is the PI on a grant from Bayer to the Broad Institute focused on the genetics and therapeutics of AF. B.M.P. serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. P.K. receives research support from the European Union, the British Heart Foundation, the Leducq Foundation, the Medical Research Council (UK) and the German Centre for Cardiovascular Research, and from several drug and device companies active in AF, and has received honoraria from several such companies. P.K. is also listed as an inventor on two patents held by University of Birmingham (Atrial Fibrillation Therapy WO 2015140571, Markers for Atrial Fibrillation WO 2016012783). K.L. is an employee of Bayer. The genotyping of participants in the Broad AF study and the expression analysis of LA tissue samples were supported by a grant from Bayer to the Broad Institute. S.N. is a consultant to Biosense Webster, Siemens and Cardiosolv. S.N. also receives research grants from NIH/NHLBI, Siemens, Biosense Webster and Imricor. S. Kathiresan has received grant support from Bayer and Amarin; holds equity in San Therapeutics and Catabasis; and has received personal fees for participation in scientific advisory boards for Catabasis, Regeneron Genetics Center, Merck, Celera, Genomics PLC, Corvidia Therapeutics and Novo Ventures. S. Kathiresan also received personal fees for consulting services from Novartis, AstraZeneca, Alnylam, Eli Lilly Company, Leerink Partners, Merck, Noble Insights, Bayer, Ionis Pharmaceuticals, Novo Ventures, Haug Partners LLC and Genetic Modifiers Newco, Inc. S.A.L. receives sponsored research support from Bristol Myers Squibb, Bayer, Biotronik and Boehringer Ingelheim, and has consulted for St. Jude Medical/Abbott and Quest Diagnostics. The remaining authors have no disclosures.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated Supplementary Information
Supplementary Figure 1 Quantile–quantile plot of combined ancestry meta-analysis.
Quantile–quantile plot of combined ancestry meta-analysis for n = 12,149,979 included variants and λGC = 1.0948.
Supplementary Figure 2 Venn diagram for genes near sentinel variants from combined ancestry meta-analysis within enriched gene sets, by functional groups.
The Venn diagram shows genes that are within enriched gene set from the gene set enrichment analysis and within 500 kb of a sentinel variant. The genes were manually grouped into functional categories based on their corresponding gene sets. The diagram shows the overlap between the genes and the functional categories.
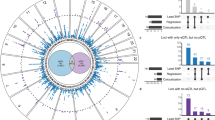
Supplementary Figure 3 Manhattan plot of European-ancestry meta-analysis.
The plot shows novel (red and purple) and known (blue) genetic loci associated with AF at a significance level of P < 1 × 10–8 (dotted line) for the European-ancestry meta-analysis (n = 537,409). The significance level accounts for multiple testing of independent variants with MAF ≥0.1% using a Bonferroni correction. P values (two-sided) were derived from a meta-analysis using a fixed-effects model with an inverse-variance-weighted approach. Loci in purple did not reach genome-wide significance in the combined ancestry meta-analysis. Gene labels correspond to the nearest gene(s). The y axis has a break between –log10 (P) of 25 and 400 to emphasize the novel loci.
Supplementary Figure 4 Quantile–quantile plot of European-ancestry meta-analysis.
Quantile–quantile plot of European-ancestry meta-analysis for n = 9,362,422 included variants and λGC = 1.1194.
Supplementary Figure 5 Manhattan plot of African-American meta-analysis.
The plot shows known (blue) genetic loci associated with AF at a significance level of P < 1 × 10–8 (dotted line), for the African-American-ancestry meta-analysis (n = 8,967). The significance level accounts for multiple testing of independent variants with MAF ≥ 0.1% using a Bonferroni correction. P values (two-sided) were derived from a meta-analysis using a fixed-effects model with an inverse-variance-weighted approach. The gene label corresponds to the nearest gene.
Supplementary Figure 6 Quantile–quantile plot of African-American-ancestry meta-analysis.
Quantile–quantile plot of African-American-ancestry meta-analysis for n = 8,640,046 included variants and λGC = 0.997.
Supplementary Figure 7 Regional plots for 4q25 for European, Japanese and African American ancestry and pairwise LD between sentinel variants at 4q25.
a–c, Regional plots of 4q25 for European-ancestry results (a, n = 537,409), Japanese-ancestry results (b, n = 36,792) and African-American-ancestry results (c, n = 8,967). LD is shown based on the 1000 Genomes phase 1 v3 reference, using the populations EUR (a), ASN (b) and AFR (c). d, Pairwise LD (r2) for the sentinel variants based on the LD from the 1000 Genomes phase 1 v3 reference for EUR (n = 379), ASN (n = 286) and AFR (n = 246) ancestry. 1000G, 1000 Genomes; AA, African American; AFR, African; ASN, Asian; EUR, European; JAP, Japanese; LD, linkage disequilibrium.
Supplementary Figure 8 Forest plots of odds ratios, and allele frequency plots, by ancestry for sentinel variants with significant heterogeneity.
a–c, Forest plots of odds ratios and pie charts of allele frequencies across ancestries (EUR, n = 537,409; JAP, n = 36,792; AA, n = 8,967; BRAZ, n = 1,664; HISP, n = 3,358) for sentinel variants with significant heterogeneity, close to PITX2 (a), NEURL (b) and ZFHX3 (c). Shown are odds ratios with 95% confidence intervals. Frequencies of the effect allele are depicted in blue; frequencies of the reference allele are depicted in orange. AA, African American; BRAZ, Brazilian; EUR, European; HISP, Hispanic; JAP, Japanese.
Supplementary Figure 9 Enrichment of atrial fibrillation–associated loci across ChromHMM regulatory regions.
a,b, Percent overlap of loci with regulatory regions (promoter, enhancer, DNase) based on the Roadmap Epigenomics Consortium 25-state model across all tissues (a) and cardiac tissues (b). Each locus includes sentinel variant and proxies with r2 > 0.6. The P values were derived from one-tailed permutation tests (n = 1,000). 1000 Genomes control loci were matched to atrial fibrillation sentinel SNPs via SNPSnap (n = 93,000). Atrial fibrillation–associated loci are from the combined ancestry analysis (n = 93). The sentinel SNP for one AF locus could not be matched in SNPSnap and was excluded from this analysis. The box plot depicts the following values: the center represents the median, the top and bottom of the box represent the first and third quartile, the whiskers reach to 1.5 times the interquartile range, and data points outside the whiskers are plotted as outliers. *P = 0.001. 1000G, 1000 Genomes; AF, atrial fibrillation.
Supplementary information
Supplementary Text, Figures and Tables
Supplementary Figures 1–9, Supplementary Tables 1, 2, 6, 8, 10 and 19–22, and Supplementary Notes
Supplementary Table 3
Known loci in combined ancestry meta-analysis
Supplementary Table 4
Gene set enrichment analysis results for combined ancestry meta-analysis
Supplementary Table 5
Novel and known loci in ancestry-specific meta-analyses
Supplementary Table 7
Loci with multiple signals identified by conditional and joint analysis for European-ancestry meta-analysis
Supplementary Table 9
Chromatin states for sentinel variants and proxies from Roadmap Epigenomics across all tissues and heart
Supplementary Table 11
Significant cis-eQTLs for sentinel variants from combined ancestry meta-analysis in GTEx heart tissues
Supplementary Table 12
Probable AF susceptibility genes for loci from combined ancestry meta-analysis
Supplementary Table 13
Transcriptome-wide results based on summary-level data from combined ancestry meta-analysis
Supplementary Table 14
Association to diseases and traits in NHGRI-EBI GWAS catalog for sentinel variants or proxies from combined ancestry meta-analysis
Supplementary Table 15
PheWAS results in UK Biobank for sentinel variants from combined ancestry meta-analysis
Supplementary Table 16
134 loci associated with atrial fibrillation
Supplementary Table 17
Baseline summary for GWAS
Supplementary Table 18
GWAS summary on genotyping, QC, imputation and analysis per study
Rights and permissions
About this article
Cite this article
Roselli, C., Chaffin, M.D., Weng, LC. et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 50, 1225–1233 (2018). https://doi.org/10.1038/s41588-018-0133-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0133-9
This article is cited by
-
Associations of combined polygenic risk score and glycemic status with atrial fibrillation, coronary artery disease and ischemic stroke
Cardiovascular Diabetology (2024)
-
Sex-specific genetic architecture of blood pressure
Nature Medicine (2024)
-
Stem cell models of inherited arrhythmias
Nature Cardiovascular Research (2024)
-
Shared genetic architecture and causal relationship between sleep behaviors and lifespan
Translational Psychiatry (2024)
-
Die Genetik von Vorhofflimmern – auf dem Weg in die Präzisionsmedizin
Herzschrittmachertherapie + Elektrophysiologie (2024)