Abstract
Many primary-tumor subregions have low levels of molecular oxygen, termed hypoxia. Hypoxic tumors are at elevated risk for local failure and distant metastasis, but the molecular hallmarks of tumor hypoxia remain poorly defined. To fill this gap, we quantified hypoxia in 8,006 tumors across 19 tumor types. In ten tumor types, hypoxia was associated with elevated genomic instability. In all 19 tumor types, hypoxic tumors exhibited characteristic driver-mutation signatures. We observed widespread hypoxia-associated dysregulation of microRNAs (miRNAs) across cancers and functionally validated miR-133a-3p as a hypoxia-modulated miRNA. In localized prostate cancer, hypoxia was associated with elevated rates of chromothripsis, allelic loss of PTEN and shorter telomeres. These associations are particularly enriched in polyclonal tumors, representing a constellation of features resembling tumor nimbosus, an aggressive cellular phenotype. Overall, this work establishes that tumor hypoxia may drive aggressive molecular features across cancers and shape the clinical trajectory of individual tumors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw sequencing data have been deposited in the European Genome-phenome Archive under accession code EGAS00001000900. Processed variant calls are available through the ICGC Data Portal under the project PRAD-CA. TCGA data are available at https://portal.gdc.cancer.gov/projects/TCGA-PRAD. Previously published CPC-GENE data are available at the European Genome-phenome Archive under accession code EGAS00001000900. Previously published CPC-GENE mRNA abundance data are available at the Gene Expression Omnibus under accession code GSE84043.
References
Wilson, W. R. & Hay, M. P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 11, 393–410 (2011).
Harris, A. L. Hypoxia: a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002).
Bristow, R. G. & Hill, R. P. Hypoxia and metabolism: hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 8, 180–192 (2008).
Weber, C. E. & Kuo, P. C. The tumor microenvironment. Surg. Oncol. 21, 172–177 (2012).
Blagosklonny, M. V. Antiangiogenic therapy and tumor progression. Cancer Cell. 5, 13–17 (2004).
Vaupel, P., Thews, O. & Hoeckel, M. Treatment resistance of solid tumors. Med. Oncol. 18, 243–260 (2001).
Dhani, N., Fyles, A., Hedley, D. & Milosevic, M. The clinical significance of hypoxia in human cancers. Semin. Nucl. Med. 45, 110–121 (2015).
Brown, J. M. & Wilson, W. R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447 (2004).
Zannella, V. E. et al. Reprogramming metabolism with metformin improves tumor oxygenation and radiotherapy response. Clin. Cancer Res. 19, 6741–6750 (2013).
Mucaj, V., Shay, J. E. S. & Simon, M. C. Effects of hypoxia and HIFs on cancer metabolism. Int. J. Hematol. 95, 464–470 (2012).
Luoto, K. R., Kumareswaran, R. & Bristow, R. G. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 4, 5 (2013).
Brizel, D. M., Dodge, R. K., Clough, R. W. & Dewhirst, M. W. Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiother. Oncol. 53, 113–117 (1999).
Nordsmark, M. & Overgaard, J. Tumor hypoxia is independent of hemoglobin and prognostic for loco-regional tumor control after primary radiotherapy in advanced head and neck cancer. Acta Oncol. 43, 396–403 (2004).
Noman, M. Z. et al. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 309, C569–C579 (2015).
Mohyeldin, A., Garzón-Muvdi, T. & Quiñones-Hinojosa, A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 7, 150–161 (2010).
Eliasson, P. & Jönsson, J.-I. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J. Cell Physiol. 222, 17–22 (2010).
Lin, P.-Y. et al. Expression of hypoxia-inducible factor-1α is significantly associated with the progression and prognosis of oral squamous cell carcinomas in Taiwan. J. Oral Pathol. Med. 37, 18–25 (2007).
Rankin, E. B. & Giaccia, A. J. Hypoxic control of metastasis. Science 352, 175–180 (2016).
Gilkes, D. M. & Semenza, G. L. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 9, 1623–1636 (2013).
Zhong, H. et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59, 5830–5835 (1999).
Graeber, T. G. et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379, 88–91 (1996).
Greijer, A. E. & van der Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 57, 1009–1014 (2004).
Bindra, R. S. et al. Down-regulation of rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell Biol. 24, 8504–8518 (2004).
Mihaylova, V. T. et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell Biol. 23, 3265–3273 (2003).
Koshiji, M. et al. HIf-1α induces genetic instability by transcriptionally downregulating mutsα expression. Mol. Cell 17, 793–803 (2005).
Lalonde, E. et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 15, 1521–1532 (2014).
Lalonde, E. et al. Translating a prognostic dna genomic classifier into the clinic: retrospective validation in 563 localized prostate tumors. Eur. Urol. 72, 22–31 (2017).
Janssens, G. O. et al. Accelerated radiotherapy with carbogen and nicotinamide for laryngeal cancer: results of a phase III randomized trial. J. Clin. Oncol. 30, 1777–1783 (2012).
Hoskin, P. J., Rojas, A. M., Bentzen, S. M. & Saunders, M. I. Radiotherapy with concurrent carbogen and nicotinamide in bladder carcinoma. J. Clin. Oncol. 28, 4912–4918 (2010).
Buffa, F. M., Harris, A. L., West, C. M. & Miller, C. J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer 102, 428–435 (2010).
Winter, S. C. et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 67, 3441–3449 (2007).
Ragnum, H. B. et al. The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br. J. Cancer 112, 382–390 (2015).
Eustace, A. et al. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin. Cancer Res. 19, 4879–4888 (2013).
Sørensen, B. S., Toustrup, K., Horsman, M. R., Overgaard, J. & Alsner, J. Identifying pH independent hypoxia induced genes in human squamous cell carcinomas in vitro. Acta Oncol. (Madr.) 49, 895–905 (2010).
Elvidge, G. P. et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition. J. Biol. Chem. 281, 15215–15226 (2006).
Hu, Z. et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 7, 9 (2009).
Seigneuric, R. et al. Impact of supervised gene signatures of early hypoxia on patient survival. Radiother. Oncol. 83, 374–382 (2007).
Hoskin, P. J. et al. Carbogen and nicotinamide in the treatment of bladder cancer with radical radiotherapy. Br. J. Cancer 76, 260–263 (1997).
Milholland, B., Auton, A., Suh, Y. & Vijg, J. Age-related somatic mutations in the cancer genome. Oncotarget 6, 24627–24635 (2015).
Yuan, Y. et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell 29, 711–722 (2016).
Li, C. H., Haider, S., Shiah, Y.-J., Thai, K. & Boutros, P. C. Sex differences in cancer driver genes and biomarkers. Cancer Res. 78, 5527–5537 (2018).
Hieronymus, H. et al. Copy number alteration burden predicts prostate cancer relapse. Proc. Natl Acad. Sci. USA 111, 11139–11144 (2014).
Vollan, H. K. M. et al. A tumor DNA complex aberration index is an independent predictor of survival in breast and ovarian cancer. Mol. Oncol. 9, 115–127 (2015).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Tan, E. Y. et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br. J. Cancer 100, 405–411 (2009).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Huang, X. et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 35, 856–867 (2009).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Sakamuro, D., Elliott, K. J., Wechsler-Reya, R. & Prendergast, G. C. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Genet. 14, 69–77 (1996).
Edwards, Y. H., Putt, W., Fox, M. & Ives, J. H. A novel human phosphoglucomutase (pgm5) maps to the centromeric region of chromosome 9. Genomics 30, 350–353 (1995).
Milosevic, M. et al. Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin. Cancer Res. 18, 2108–2114 (2012).
Fraser, M. et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 (2017).
Espiritu, S. M. G. et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 173, 1003–1013 (2018).
Hopkins, J. F. et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 8, 656 (2017).
Jackson, W. C. et al. Intermediate endpoints after postprostatectomy radiotherapy: 5-year distant metastasis to predict overall survival. Eur. Urol. https://doi.org/10.1016/j.eururo.2017.12.023 (2018).
Chua, M. L. K. et al. A prostate cancer “nimbosus”: genomic instability and schlap1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur. Urol. 71, 183–192 (2017).
Chua, M. L. K., van der Kwast, T. H. & Bristow, R. G. Intraductal carcinoma of the prostate: anonymous to ominous. Eur. Urol. 67, 496–503 (2017).
Benita, Y. et al. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 37, 4587–4602 (2009).
Ding, Z. et al. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 42, e75 (2014).
Semenza, G. L. Hypoxia, clonal selection, and the role of hif-1 in tumor progression. Crit. Rev. Biochem. Mol. Biol. 35, 71–103 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Haider, S. et al. Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia. Genome. Biol. 17, 140 (2016).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Preprint at https://www.biorxiv.org/content/early/2017/08/30/161562 (2017).
Zundel, W. et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 14, 391–396 (2000).
Hong, M. K. H. et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 6, 6605 (2015).
Mertins, P. et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 (2016).
Zhang, H. et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 (2016).
Zhang, B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 (2014).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Irizarry, R. A. et al. Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 31, e15 (2003).
Fisher, S. et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation dna sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Govind, S. K. et al. ShatterProof: operational detection and quantification of chromothripsis. BMC Bioinformatics 5, 78 (2014).
Masella, A. P. et al. BAMQL: a query language for extracting reads from BAM files. BMC Bioinformatics 17, 305 (2016).
Ha, G. et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 24, 1881–1893 (2014).
Deshwar, A. G. et al. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 16, 35 (2015).
Parker, C. et al. Polarographic electrode study of tumor oxygenation in clinically localized prostate cancer. Int. J. Radiat. Oncol. 58, 750–757 (2004).
P’ng, C. et al. BPG: seamless, automated and interactive visualization of scientific data. Preprint at https://www.biorxiv.org/content/early/2017/06/26/156067 (2017).
Reimand, J., Kull, M., Peterson, H., Hansen, J. & Vilo, J. g:Profiler: a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007).
Acknowledgements
This study was conducted with the support of Movember funds through Prostate Cancer Canada, and with the additional support of the Ontario Institute for Cancer Research, funded by the Government of Ontario. This work was supported by Prostate Cancer Canada and is funded by the Movember Foundation (grant RS2014-01). P.C.B. was supported by a Terry Fox Research Institute New Investigator Award, a Prostate Cancer Canada Rising Star Fellowship, and a Canadian Institutes of Health Research (CIHR) New Investigator Award. This work has been funded by Fellowships from the CIHR and the Ontario government to V.B. and E.L. S.K.L. is supported as a Movember Rising Star award recipient funded by the Movember Foundation (grants RS2014-03, D2015-12 and D2017-1811), the Telus Motorcycle Ride For Dad (Huronia Branch) and a Ministry of Research and Innovation Early Researcher Award. The authors thank the Princess Margaret Cancer Centre Foundation and Radiation Medicine Program Academic Enrichment Fund for support (to R.G.B.). This work was supported by a Terry Fox Research Institute Program Project Grant. R.G.B. is supported as a recipient of a Canadian Cancer Society Research Scientist Award. Laboratory work for R.G.B is supported by the CRUK Manchester Institute through Cancer Research UK. The authors thank all members of the Boutros and Bristow laboratoriess for helpful suggestions.
Author information
Authors and Affiliations
Contributions
V.B. conducted bioinformatics and statistical analysis; V.B., L.Y.L., E.L., J.L., R.L., Y.J.S., S.M.G.E., L.E.H., F.Y., V.H., T.N.Y., C.Q.Y. and V.Y.S. performed data processing; C.H., J.R., T.V. and X.H. performed in vitro experiments; V.B. performed data visualization; M.F., M.L.K.C., T.v.d.K., S.K.L., P.C.B. and R.G.B. supervised research; V.B., P.C.B. and R.G.B. initiated the project; V.B. wrote the first draft of the manuscript; all authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
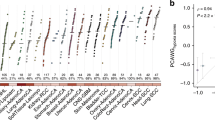
Supplementary Figure 1 Pan-cancer hypoxia assessment in primary tumors.
a,b, Tumor hypoxia scores based on the Winter and Ragnum hypoxia signatures for 7,791 independent tumors from 19 tumor types. The median hypoxia score within each cancer type is indicated by the horizontal line. Sample sizes for each tumor type are listed along the bottom (representing independent tumors). Intertumoral variability in hypoxia, measured by interquartile range (IQR), is also shown along the bottom. c, Hypoxia scores from eight independent hypoxia signatures are highly correlated. For each pair of signatures, the Spearman’s ρ for hypoxia scores was calculated for each of the 19 tumor types. The value in each box represents the median ρ for a pair of signatures (ρ = 0.42 ± 0.21, overall mean ± s.d.). d, Comparison of hypoxia rankings across signatures for 19 tumor types from TCGA. Median hypoxia scores for the 19 tumor types were scaled from –1 to 1. Squamous carcinomas of the head and neck, lung and cervix are consistently observed as the most hypoxic. e, Comparison of the distribution of hypoxia scores between the Winter, Buffa and Ragnum hypoxia scores (n = 7,791 independent tumors; Spearman’s ρ, P value calculated using algorithm AS89). Tumor type codes are defined in Supplementary Table 2.
Supplementary Figure 2 Influence of hypoxia on protein abundance.
a–c, Comparison of tumor hypoxia scores generated from mRNA abundance and protein abundance data using the Buffa hypoxia signature for BRCA (breast invasive carcinoma) (a), OV (ovarian serous cystadenocarcinoma) (b) and COADREAD (colon adenocarcinoma and rectum adenocarcinoma) (c). Protein-abundance-based hypoxia scores were significantly correlated with mRNA-abundance-based hypoxia scores for all three tumor types. d, The abundance of ten proteins was significantly correlated with protein-based hypoxia scores across all three cancers (FDR < 0.05 in all three cancer types). nBRCA = 77 independent tumors, nOV = 102 independent tumors, nCOADREAD = 86 independent tumors for a–d. Spearman’s ρ was used to determine each correlation and P values were calculated using algorithm AS89.
Supplementary Figure 3 Pan-cancer associations of hypoxia with sex, age and ancestry, and power to detect hypoxia-associated SNVs.
a–c, Significant differences in tumor hypoxia between females and males were not seen consistently based on all three hypoxia signatures for any tumor type (Mann–Whitney U test). Bonferroni-adjusted P values are shown along the top. d, Younger patients with lung adenocarcinoma had higher hypoxia scores. However, this association was confounded by smoking status. Background color indicates Bonferroni-adjusted P values (algorithm AS89) and dot size indicates the magnitude of the correlation (Spearman’s ρ). e–g, Significant differences in tumor hypoxia were consistently seen in patients with breast invasive carcinomas (Kruskal–Wallis test). Bonferroni-adjusted P values are shown along the top. h, Power analysis for a Mann–Whitney U test to detect hypoxia score differences between patients with and without an SNV. The breast invasive carcinoma (BRCA), lung adenocarcinoma (LUAD) and renal clear cell carcinoma (KIRC) cohorts were well powered for this analysis. All tumor type codes are defined in Supplementary Table 2. Tukey box plots are shown in a–c, e–g. na–c, d, e–g independent tumors = 408, 408, 391 BLCA; NA, 1,093, 997 BRCA; NA, 304, 259 CESC; 377, 377, 342 COADREAD; 159, 159, 158 GBM; 520, 519, 503 HNSC; 533, 533, 526 KIRC; 290, 287, 273 KIRP; 515, 515, 504 LGG; 371, 370, 359 LIHC; 515, 496, 448 LUAD; 501, 492, 388 LUSC; NA, 304, 290 OV; 178, 178, 174 PAAD; 179, NA, 174 PCPG; NA, 333, 143 PRAD; 469, 461, 459 SKCM; 501, 501, 409 THCA; NA, 174, 153 UCEC. All tests were two-sided.
Supplementary Figure 4 Validation of hypoxia-associated alterations in breast cancer, associations with subtypes, pan-cancer signature agreements and miRNA–protein associations.
a, Validation of hypoxia-associated CNAs and SNVs in breast cancer using an independent cohort (Nature 486, 346–352, 2012; n = 1,859 independent tumors; Mann–Whitney U test). b–d, Hypoxia scores were significantly different between subtypes of breast cancer in TCGA (b), METABRIC (c) and CPTAC (d) where hypoxia scores were calculated based on protein abundance (n = 817, 1,985 and 77 independent tumors, respectively; Kruskal–Wallis test; Tukey box plots are shown). e, Elevated protein-based hypoxia scores are associated with elevated abundance of TP53 (n = 68 independent tumors; Spearman’s ρ, AS89). f–o, Patients with mutations in TP53 had elevated hypoxia scores compared to patients with wild-type TP53 within the different breast cancer subtypes in TGGA and METABRIC (Mann–Whitney U test). p–r, We assessed agreement between the Winter, Buffa and Ragnum signatures by comparing whether the different signatures considered a driver gene to be significantly (FDR < 0.05) associated with hypoxia or not. For 98.2 ± 3.1%, 72.4 ± 23.4% and 61.9 ± 16.9% (mean ± s.d.) of driver SNVs (p), CNAs (q) and miRNAs (r), respectively, all three signatures agreed on it being associated or not associated with hypoxia, indicating that the molecular associations we highlight in Fig. 2 are broadly informative about biology across cancer types. np,q and r independent tumors = 388, 405 BLCA; 960, 753 BRCA; 190, 304 CESC; 259, 295 COADREAD; 137 GBM; 497, 478 HNSC; 431, 254 KIRC; 280, 290 KIRP; 513, 512 LGG; 360, 367 LIHC; 475, 447 LUAD; 178, 342 LUSC; 246, 288 OV; 119, 178 PAAD; 162, 179 PCPG; 333, 330 PRAD; 290, 97 SKCM; 486, 500 THCA; 8, 174 UCEC. Correlations between miR-210, a hypoxia-associated miRNA, and protein abundance in BRCA (s; n = 32 independent tumors) and OV (t; n = 139 independent tumors). Results for the top 20 proteins are shown. The protein abundance of lactate dehydrogenase A (LDHA) was significantly positively correlated with the abundance of miR-210 in BRCA (s) and OV (t) (Spearman’s ρ, algorithm AS89). Tumor type codes are defined in Supplementary Table 2. All tests were two-sided.
Supplementary Figure 5 Hypoxia associations with clinicopathological features, TERT mRNA abundance, PTEN mRNA abundance and mitochondrial mutations.
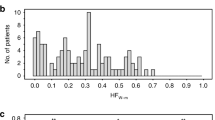
a, Tumor hypoxia increases with T-category (n = 405 independent tumors). b, Tumor hypoxia significantly differs by Gleason score (n = 479 independent tumors). c, Mutations in the origin of heavy strand replication (OHR) and MYC have previously been associated with poor prognosis in localized prostate cancer (Nat. Commun. 8, 656, 2017). In line with this prognostic association, a significant difference in hypoxia score was noted based on OHR and MYC mutation status (n = 152 independent tumors). Patients in the CPC-GENE (d; n = 191 independent tumors) and TCGA (e; n = 308 independent tumors) cohorts with the aggressive IDC-CA pathological feature have significantly higher tumor hypoxia scores compared to patients without IDC-CA. f, PTEN mRNA abundance is negatively correlated with the mRNA abundance of TERT, a HIF-1A target, in the TCGA cohort (n = 333 independent tumors; Spearman’s ρ, algorithm AS89). g, PTEN mRNA abundance levels differ based on hypoxia and TERT mRNA abundance (n = 333 independent tumors). h, Monoclonal tumors with high hypoxia scores have higher TERT mRNA abundance than other subgroups of tumors based on clonality and hypoxia (n = 125 independent tumors). i, The mRNA abundance of PTEN is modulated by both PTEN copy number status and hypoxia (n = 60 independent tumors). Tukey box plots are shown in a–e, g–i. Kruskal–Wallis tests used for a–c, g–i. Mann–Whitney U test used for d and e. All tests were two-sided.
Supplementary Figure 6 Enrichment, mRNA confirmation and pathway analysis of hypoxia-associated CNAs, hypoxia-associated events in monoclonal tumors and a model for hypoxia as a driver of aggressive prostate cancer.
a, Hypoxia-associated CNA hits were enriched on chromosomes 7 and 10. Numbers along the top are Bonferroni-adjusted P values (n = 360 independent tumors; hypergeometric test). Comparison of mRNA abundance in TCGA (b; n = 148 independent tumors) and CPC-GENE (c; n = 210 independent tumors) between tumors that have a copy number loss or gain to tumors that are neutral for hypoxia-associated genes at the CNA level (Mann–Whitney U test). FDR-adjusted P values are shown along the top (Mann–Whitney U test). Fold changes are shown along the bottom. d, Pathways related to the 20 genes associated with hypoxia at the CNA level and functionally confirmed at the mRNA level. Dot size indicates the number of genes in the data set that are in the gene set. Dot color indicates the q value. The color of the lines connecting the nodes indicates the overlap between connected gene sets. e,f, PTEN mRNA abundance is influenced by hypoxia together with PTEN copy number status (e; n = 60 independent tumors; Kruskal–Wallis test) and IDC-CA (f; n = 57 independent tumors; Kruskal–Wallis test). g, Allelic loss of PTEN and IDC/CA are often observed in the same monoclonal tumors (Fisher’s exact test). h, Hypoxia applies a selective pressure within prostate tumors, driving the selection of aggressive tumor subclones. Driver mutations are shown as yellow, orange and red stars. The onset of hypoxia is shown as a gradient from white to blue, with subsequent reoxygenation shown as a gradient from blue to white. Once normal levels of oxygen are reestablished, aggressive subclones that survived the hypoxic microenvironment can rapidly expand. Tukey box plots are shown in b, c, e and f. All tests were two-sided.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note
Supplementary Table 1
Pancancer hypoxia scores
Supplementary Table 2
TCGA tumor type descriptions
Supplementary Table 3
Molecular correlates of hypoxia in breast, lung and kidney cancer
Supplementary Table 4
CPC-GENE data
Supplementary Table 5
Hypoxia-associated CNAs in localized prostate cancer
Supplementary Table 6
miRNA sequence information
Rights and permissions
About this article
Cite this article
Bhandari, V., Hoey, C., Liu, L.Y. et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet 51, 308–318 (2019). https://doi.org/10.1038/s41588-018-0318-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0318-2
This article is cited by
-
In-situ-sprayed therapeutic hydrogel for oxygen-actuated Janus regulation of postsurgical tumor recurrence/metastasis and wound healing
Nature Communications (2024)
-
Spatial transcriptomics reveals that metabolic characteristics define the tumor immunosuppression microenvironment via iCAF transformation in oral squamous cell carcinoma
International Journal of Oral Science (2024)
-
Novel biomarker SARIFA in colorectal cancer: highly prognostic, not genetically driven and histologic indicator of a distinct tumor biology
Cancer Gene Therapy (2024)
-
A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer
Nature Medicine (2024)
-
Metabolic heterogeneity in cancer
Nature Metabolism (2024)