Abstract
Mitochondria are essential cellular organelles that play critical roles in cancer. Here, as part of the International Cancer Genome Consortium/The Cancer Genome Atlas Pan-Cancer Analysis of Whole Genomes Consortium, which aggregated whole-genome sequencing data from 2,658 cancers across 38 tumor types, we performed a multidimensional, integrated characterization of mitochondrial genomes and related RNA sequencing data. Our analysis presents the most definitive mutational landscape of mitochondrial genomes and identifies several hypermutated cases. Truncating mutations are markedly enriched in kidney, colorectal and thyroid cancers, suggesting oncogenic effects with the activation of signaling pathways. We find frequent somatic nuclear transfers of mitochondrial DNA, some of which disrupt therapeutic target genes. Mitochondrial copy number varies greatly within and across cancers and correlates with clinical variables. Co-expression analysis highlights the function of mitochondrial genes in oxidative phosphorylation, DNA repair and the cell cycle, and shows their connections with clinically actionable genes. Our study lays a foundation for translating mitochondrial biology into clinical applications.
Similar content being viewed by others
Main
Mitochondria are crucial cellular organelles in eukaryotes, and there can be several hundred mitochondria in a single human cell1. Known as ‘the powerhouses of the cell’, mitochondria play essential roles in generating most of the cell’s energy through oxidative phosphorylation2. Despite its small size (16.6 kilobases (kb)), the circular mitochondrial genome encodes 13 proteins that form respiratory chain complexes with other proteins of nuclear origin3. The involvement of mitochondria in carcinogenesis has long been suspected4,5 because altered energy metabolism is a common feature of cancer6. Furthermore, mitochondria play important roles in other tasks, such as biosynthesis, signaling, cellular differentiation, apoptosis, maintaining control of the cell cycle and cell growth, all of which are intrinsically linked to tumorigenesis5,7.
In several recent studies, molecular characterization of mitochondria was performed in cancer by using next-generation sequencing data8,9,10,11,12,13, but these studies usually describe one specific dimension of the mitochondrial genome (for example, somatic mutations) based on relatively small sample cohorts. Furthermore, due to the whole-exome sequencing data employed, the relatively low depth of mitochondrial genomes limits the accuracy and scope of these studies. Thus, a comprehensive, multidimensional molecular portrait of mitochondria across a broad range of cancer types has not been achieved. Moreover, previous studies have focused on the patterns of mitochondrial alterations alone, without fully exploring the interplay between the mitochondrial genome and the nuclear genome, as well as the biomedical significance of mitochondrial alterations.
The Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium aggregated whole-genome sequencing (WGS) data from 2,658 cancers across 38 tumor types generated by the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) projects. These sequencing data were re-analyzed with standardized, high-accuracy pipelines to align to the human genome (reference build hs37d5) and identify germline variants and somatically acquired mutations, as described14. Meanwhile, TCGA has generated RNA sequencing (RNA-seq) data from a large number of patient samples, which allow for assessment of the transcriptional activities of mitochondrial genes15. These large-scale datasets create a tremendous resource for characterizing cancer mitochondrial genomes at an unprecedented level (Fig. 1a). We first characterized mitochondrial somatic mutations, nuclear transfers and copy numbers, then investigated their interactions with nuclear somatic alterations16,17, and finally examined the expression profiles of mitochondrial genes and their connections with clinically relevant nuclear genes.
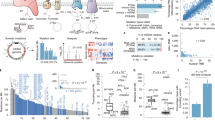
a, Overview of our multidimensional and integrated mitochondrial genome analyses. b, Landscape of mtDNA somatic substitutions. The numbers represent the mitochondrial genome coordinates. The outer (blue) circle shows the density of all variants with VAF > 1%. The inner (red) circle shows the density of variants with VAF > 3%. c, Highly consistent mtDNA mutational spectrum across 21 cancer tissue groups. Average numbers of somatic substitutions per sample are also shown (right). d, Correlation between the highest VAF of mtDNA mutations in a cancer tissue and patient age at the time of diagnosis. The correlation was based on the 2,414 patients with both age and somatic single-nucleotide variant information available. The shaded region represents the 95% confidence interval for the predictions from a linear model with the highest VAF as the response variable and patient age as the explanatory variable. e, Correlations between the numbers of nuclear and mtDNA somatic mutations. The associations were tested among samples with both nuclear and mtDNA somatic mutations available (with sample sizes labeled on plot) using Spearman’s rank correlation. Magenta bars indicate significant positive correlations (P < 0.05). Asterisks indicate that both nuclear and mitochondrial somatic mutations were correlated with patient age in that cancer type (P < 0.05). f, Proportions of tumor samples harboring different categories of somatic alterations: nuclear driver alterations only (red); both nuclear driver alterations and mtDNA mutations (VAF > 10%, green); and mtDNA mutations without known nuclear drivers (blue). AML, acute myeloid leukemia; BNHL, B cell non-Hodgkin lymphoma; CA, carcinoma; ChRCC, chromophobe renal cell carcinoma; CLL, chronic lymphocytic leukemia; CNS, central nervous system; eso, esophageal; GBM, glioblastoma; HCC, hepatocellular carcinoma; leiomyo, leiomyosarcoma; liposarc, liposarcoma; medullo, medulloblastoma; MPN, myeloproliferative neoplasm; oligo, oligometastatic; osteosarc, osteosarcoma; PiloAstro, pilocytic astrocytoma; RCC, renal cell carcinoma; SCC, squamous cell carcinoma; TCC, transitional cell carcinoma; adenoCA, adenocarcinoma; epith, epithelioid.
Results
Mutational landscape of cancer mitochondrial genomes
To characterize somatic mutations in mitochondrial genomes across cancer types, we extracted the mitochondrial DNA (mtDNA) mapped reads of 2,658 cancer and matched control sample pairs from the PCAWG Consortium. The samples we surveyed covered 21 cancer tissues and 38 specific cancer types (Supplementary Table 1). On average, the sequencing depth for the mitochondrial genome was 9,959×, which was much higher than that obtained from whole-exome sequencing data, allowing for confident detection of somatic mutations at a very low heteroplasmic level (variant allele fraction (VAF) > 1%; Supplementary Fig. 1). By applying a well-designed computational pipeline that carefully considered various potentially confounding factors (for example, sample cross-contamination, mismapping of reads from nuclear mtDNA-like sequence18, and artifactual mutations caused by oxidative DNA damage during library preparation19), we identified a total of 7,611 somatic substitutions and 930 small indels in 2,536 high-quality cancer samples (122 samples were excluded in the mutation analysis for the issues mentioned above; Supplementary Fig. 2 and Methods). The high reliability of the mutations was confirmed by long-range PCR-based validation (Supplementary Table 2) and by inspection of the mutational spectrum of the very low-VAF mutation candidates (Supplementary Fig. 3).
Of the 7,611 substitutions, >85% were clearly heteroplasmic, showing VAFs lower than 0.6 (average: 0.2; median: 0.045). Overall, mtDNA mutations located in the transcribed regions were also found in RNA-seq with similar VAFs, except for a fraction of transfer RNA (tRNA) mutations showing much higher VAFs in transcripts due to the accumulation of unprocessed tRNA precursors during the processing of polycistronic mitochondrial transcripts10 (Supplementary Fig. 4). Across all of the cancer samples, we observed several mutational hotspots in the regulatory D-loop region and the ND4 gene (Fig. 1b). Of the 13 protein-coding genes, ND5 was the most frequently mutated in most cancer types, while ND4 was most frequently mutated in prostate and lung cancers, and COX1 was most frequently mutated in breast, cervical and bladder cancers (Supplementary Fig. 5). We identified that cancer type and gene identity were associated with the mutation status of the 13 coding genes (log-linear model, Pcancer type < 2.2 × 10−16; Pgene < 2.2 × 10−16), but the effect of their interaction was not significant (Pcancer type × gene = 0.12).
In contrast with somatic mutations in nuclear genomes (where cancer type-specific mutational signatures are observed)20, mtDNA mutational signatures were very similar across tumor types, with C:G>T:A (58.3%) and T:A>C:G (34.2%) substitutions being the most and second most frequent mutation types, respectively (Fig. 1c and Supplementary Fig. 6). Indeed, the impact of well-known carcinogens (for example, tobacco smoking (C:G>A:T dominant; signature 4), ultraviolet light (C:G>T:A dominant at dipyrimidine contexts; signature 7) and reactive oxygen species (G:C>T:A dominant)) were minimal (Supplementary Fig. 7) even in lung and skin cancers (the latest mutational signatures of nuclear genomes are available from the Catalogue of Somatic Mutations in Cancer database: https://cancer.sanger.ac.uk/cosmic/signatures). Instead, the vast majority of mtDNA mutations were manifesting extreme replicational mtDNA strand bias9,21,22; that is, predominant G>A and T>C substitutions and deficient complementary C>T and A>G substitutions on the light (L) strand of the mtDNA genome sequence (+strand of the revised Cambridge Reference Sequence) despite the relative depletion of guanines and thymines on the L strand (Supplementary Fig. 6). These mutational signatures suggest that mitochondria-specific, replication-coupled mutational processes (such as mtDNA polymerase gamma error9,21,23 or other replication-coupled DNA damage mechanisms) are dominantly responsible for somatic mtDNA mutations in cancer.
In agreement with its endogenous origin, we observed clock-like properties in mtDNA mutations as nuclear genome mutations24. The number of mtDNA mutations in our study was largely proportional to the age of the patient at the time of tissue sampling (Supplementary Fig. 8). In addition, the maximum VAF of somatic mutations in a cancer tissue showed positive correlation with age, on average (Fig. 1d; P < 2.2 × 10−16). Collectively, these results suggest that the vast majority of mtDNA somatic mutations were: (1) acquired at an earlier age when the cell lineage was phenotypically normal; and (2) overall shifted towards homoplasmy throughout life in the cellular lineage of the neoplastic cells. The spread to homoplasmy can, in theory, be caused by either physiological advantage (selection) or a series of asymmetric segregations during cell divisions (drift)25, or both.
To further assess the potential impact of mtDNA mutations, we performed integrative analysis by examining alterations from mitochondrial and nuclear genomes simultaneously17. We observed significantly positive correlations between the mutation burdens of mitochondrial and nuclear genomes in several cancer types, with the highest correlations observed in kidney chromophobe and thyroid cancers (magenta bars in Fig. 1e). Some of these correlations may be explained by the age effect, as the mutation numbers in both mitochondrial and nuclear genomes were significantly correlated with patient age in the corresponding cancer types (bars marked with an asterisk in Fig. 1e). In addition, we examined the mtDNA mutation frequency in the context of nuclear drivers. Although nuclear driver alterations exist in the majority of patients in most cancer types, a notable proportion of patients (22.2% with kidney chromophobe cancer and 18.8% with thyroid cancer) bear non-silent mtDNA mutations but no known nuclear drivers, suggesting a potential functional contribution of mtDNA mutations in the absence of nuclear drivers in these cancer types (Fig. 1f).
Hypermutation process in mitochondrial genomes
Hypermutation processes have been well established for a small proportion of cancer nuclear genomes (for example, microsatellite instability)26,27, but have not been reported for mitochondrial genomes. Of the 2,536 cancer samples surveyed, seven cases showed extremely large numbers of mtDNA somatic substitutions (>13 mutations), which were larger than expected from the background distribution (Fig. 2a; around three somatic substitutions per sample on average, with a standard deviation of 2.6). The mutational spectra in these hypermutated samples were sometimes clearly distinguished from the background L-strand G>A and T>C substitution dominant signature (Fig. 2b), suggesting that the massive numbers of mutations are not the consequence of the gradual accumulation of ordinary mtDNA substitutions.
a, Distribution of mtDNA mutations. The blue curve represents the calculated ratio between observed and expected numbers of samples for each bin (right-hand y axis). b, Mutational spectrum of the seven hypermutated mitochondrial genomes identified. The P values were generated by chi-squared test without multiple comparison adjustment (**P < 0.01; ****P < 0.0001). c, Distribution of the 33 somatic mutations in the breast cancer sample of mtDNA hypermutation (sample ID: SP6730). d, Proposed model of the mtDNA hypermutation process in SP6730. rRNAs, ribosomal RNAs.
The most striking case was a breast cancer sample (sample ID: SP6730) harboring 33 mutations, 30 of which were localized in a 2-kb region (Fig. 2c), resulting in a local hypermutational rate (>75× higher than the background mutational rate). The mutations were neither of germline origin (~70% were novel) nor caused by sequencing errors, as confirmed by independent exome and RNA-seq analyses (Supplementary Fig. 9). Interestingly, most of the localized mutations (n = 28) were T>C substitutions on the L strand (Fig. 2b,c) and were co-clonal of each other, with highly similar VAFs (~7%) and direct physical phasing by Illumina sequencing reads (Supplementary Fig. 9). Collectively, these lines of evidence strongly suggest that the 28 localized mutations (19 missense, four silent and five tRNA mutations) were acquired by a ‘single-hit’ catastrophic mutational mechanism with strand-specific T>C substitutions as a dominant spectrum, reminiscent of the kataegis phenomenon in the nuclear genome28 (Fig. 2c) and/or complex somatic mutations reported in mtDNA29. The mutated mtDNA copy is then likely to shift to appreciable VAF (~7% frequency) by a series of replications throughout the cell lineages, despite the low probability of causation of a defective phenotype.
Cancer type-specific selective pressures on mtDNA mutations
To investigate the functional consequences of mtDNA genes, we examined the dN/dS ratio (a common measure of selective pressure on missense mutations) with consideration of the unique mtDNA mutational signature9. We found that dN/dS was overall close to 1 for missense mutations at different VAFs across cancer types, suggesting that overall selection for mtDNA missense mutations is nearly neutral (Supplementary Fig. 10). However, it should not be interpreted that all missense mtDNA mutations are passengers.
For truncating mutations on the 13 mtDNA genes, we found evidence of negative selection in most cancer types, suggesting the importance of intact mitochondrial function in cancer cells. For example, the VAFs of mtDNA truncating mutations were notably more suppressed than those of missense or silent mutations (Fig. 3a). Interestingly, kidney, colorectal and thyroid cancers showed the opposite trend, where mtDNA truncating mutations exhibited significantly higher VAFs than the background (F-test, P < 2.2 × 10−16; Fig. 3a). The enrichment of nearly homoplasmic (>60% VAF) truncating mutations was very striking in kidney cancers, especially in chromophobe and papillary types, suggesting that the inactivation of the normal mitochondrial function is an important step in tumorigenesis30 (Fig. 3b and Supplementary Fig. 11). The mtDNA truncating mutations were enriched in ND5. Compared with kidney chromophobe and colorectal cancers, kidney papillary cancers harbored ND5 truncation mutations enriched in the amino-terminal region (Fisher’s exact test, P = 0.05; Fig. 3c). Integrating with the mutation data of nuclear genes, we found that the high VAF truncating mutations in the two kidney cancer types were mutually exclusive to the mutations of known cancer genes (Fisher’s exact test, P = 0.01; Fig. 3d). Moreover, samples with mtDNA truncating mutations showed upregulation of gene expression in cancer-related pathways, such as mammalian target of rapamycin signaling, tumor necrosis factor-α signaling, oxidative phosphorylation and protein secretion (false discovery rate (FDR) < 0.05; Fig. 3e). Collectively, these results strongly suggest functional oncogenic impacts of mitochondrial truncating mutations in the initiation and clonal evolution of the specific cancer types.
a, Distinct VAF accumulation curves of truncating mutations between kidney/colorectal/thyroid cancers and other cancer types. For comparison, similar curves were generated for silent and missense mutations, which are overall functionally neutral, in other types of cancer after normalization of mutation numbers. Generally, fewer truncating mutations were observed at higher allele-frequency levels (red), except for kidney, colorectal and thyroid cancer types (blue). n = number of samples. b, Kidney chromophobe, kidney papillary, colorectal and thyroid cancers accumulated excessive high-allele-frequency truncating mutations (sample sizes in parentheses). Areas under the curve across cancer types for a VAF interval of 0.6–1.0 (from Supplementary Fig. 11a); were calculated and are shown. Their distribution is shown by a box plot. The boundaries of the box mark the first and third quartile, with the median in the center, and whiskers extending to 1.5× the interquartile range from the boundaries. c, Distribution patterns of truncating mutations in ND5. C, carboxy; N, amino; Proton_antipo_N, NADH-ubiquinone oxidoreductase (complex I), chain 5 N-terminus; Proton_antipo_M, proton-conducting membrane transporter; NADH5_C, NADH dehydrogenase subunit 5 C terminus. d, Heat map of mtDNA truncating mutations with recurrent somatic mutations in cancer genes in kidney chromophobe and kidney papillary cancers. MT_truncating standas for mitochondrial truncating mutations, which include frameshift mutations and stop-gain mutations. Statistical significance of mutual exclusivity between mutations was assessed by Fisher’s exact test. SNV, single-nucleotide variant. e, Heat map of the signaling pathways enriched by nuclear genes upregulated in cancer samples with truncating mutations. A dot indicates FDR < 0.05. IFN-γ, interferon-γ; IL-6, interleukin-6; JAK, Janus kinase; mTORC1, mammalian target of rapamycin complex 1; NF-κB, nuclear factor κB; STAT3, signal transducers and activators of transcription 3; TNF-α, tumor necrosis factor-α; KRAS, KRAS proto-oncogene, GTPase.
Somatic transfer of mtDNA into the nuclear genome
The migration of mtDNA into the nuclear genome has been assessed using different technologies31,32,33. Recently, somatic mtDNA nuclear transfers (SMNTs) have been more systematically studied in nucleotide resolution11, mostly in breast cancers. In this study, of the 2,658 cancer cases across 21 tissue types, we found 55 positive cases (2.1% overall positive rate) (Methods). The SMNT rate varied according to the cancer tissue type (Fisher’s exact test, P < 1 × 10−5; Fig. 4a). For example, lung, skin, breast and uterine cancers showed frequencies higher than 5%. In particular, human epidermal growth factor receptor 2-positive (HER2+) breast cancers and squamous cell lung cancers showed positive rates of 16.0% (four out of 25 cases) and 14.6% (seven out of 48 cases), respectively, which were significantly higher than the average (Fisher’s exact test, P < 0.003 and P < 0.001, respectively). In contrast, we did not find any positive cases from blood, kidney, esophagogastric, liver, prostate and colorectal cancers. The samples with SMNTs showed a much higher number of global and local structural variations in the nuclear genome than the control samples16 (P = 1 × 10−4; Fig. 4b and Supplementary Fig. 12). SMNT integration sites (breakpoints) were spatially closer to inversion and translocation breakpoints than expected (Fig. 4c). These results suggest that the integration of mtDNA segments into nuclear DNA is often mechanistically combined with some specific processes underlying structural variations in the nuclear genome.
a, Frequency of SMNTs in different cancer tissues. Circle size indicates the sample size of a given cancer type. ER, estrogen receptor. b, Numbers of structural variant breakpoints in samples with and without SMNTs. Sample sizes are labeled below the boxes. The P values (***P < 0.001; ****P < 0.0001) were generated by two-sided t-test without multiple comparison adjustment. c, Distances from SMNT breakpoints to the nearest structural variant breakpoints are shorter than random expectation for all and each type of structural variant. Sample sizes are labeled in the centers of the boxes. The P values (**P < 0.01; ***P < 0.001) were generated by two-sided t-test without multiple comparison adjustment. bp, base pair; exp, expected; Mb, megabase; NS, not significant; obs, observed. d, Circos plot of three independent SMNT events in a bladder cancer genome (sample ID: SP953), showing 23 human chromosomes in the outer layer, as well as copy numbers of nuclear cancer genomes (inner layer; black dots); chromosomal rearrangements (gray curves) and SMNTs (red curves). A summary of three SMNTs with genomic coordinates in numbers is depicted below the Circos plot with breakpoints. e, An SMNT event found in a HER2+ breast cancer genome (sample ID: SP10563), leading to a tandem duplication process of ERBB2 exons 10–23 and their subsequent expression. The novel exon junction is supported by the RNA reads from the corresponding RNA-seq data. In all boxplots, the boundaries of the boxes mark the first and third quartiles, with the median in the center, and whiskers extending to 1.5× the interquartile range from the boundaries.
Despite the overall low SMNT frequency (~2%), some cancer samples showed up to three independent SMNT events (Fig. 4d and Supplementary Fig. 13). Sometimes, somatically transferred mtDNA segments were extensively rearranged (Supplementary Fig. 13b), implying extreme genomic instability at the time of the SMNT events. We observed 42 SMNT events in 35 tumor cases that were integrated in the middle of genes (n = 42), mostly in introns (n = 37), with a few events in the protein-coding regions (n = 3) and in the untranslated regions (n = 2) (Supplementary Table 3). Among these, open reading frames of at least 23 genes (23/42 = 55%), including cancer genes such as ERBB2, FOLH1 and ULK2, were predicted to be altered by these SMNTs and their combined structural variant events in the vicinity (Supplementary Fig. 14). Of particular interest, one SMNT was involved in transforming focal amplification of the ERBB2 gene in a HER2+ breast cancer genome (Fig. 4e).
Copy-number and structural variations of mtDNA
Although previous studies have examined mtDNA copy numbers in individual cancer types34,35,36 or from a collection of whole-exome sequencing data12, we performed a systematic and accurate analysis of mtDNA copy numbers per cell over the largest sample cohort with WGS data so far, with consideration to confounding factors such as the normal-cell contamination and genome ploidy of tumor cells (Supplementary Fig. 15 and Methods).
Based on the 2,157 cancer samples that passed the purity filter, we observed great variation in mtDNA copy numbers across and within cancer types: mtDNAs were most abundant in samples of ovarian cancer (median: 644 copies per cell) and least abundant in myeloid cancer (median: 90 copies per cell) (Fig. 5a). Different cancer subtypes originating from the same tissue sometimes showed distinct mtDNA copy-number distributions (Fig. 5b and Supplementary Fig. 16). For example, the mtDNA copy numbers for kidney chromophobe were significantly higher than those for kidney clear cell and kidney papillary (analysis of variance (ANOVA), P < 7.8 × 10−6; Fig. 5b). This may be interlinked with the general inadequacy of mitochondrial quality control and resultant increase in the steady-state mtDNA copy number, as seen in renal oncocytoma37. Indeed, we found that the mtDNA copy number was significantly higher in the samples with high-allele-frequency truncating mutations (ANOVA with consideration of confounders, P < 1.7 × 10−4; Fig. 5c), suggesting that the dosage effect of mtDNAs was selected to compensate for the deleterious effect of truncating mutations. For the cancer samples with WGS data from matched normal tissues (n = 507), we observed increased mtDNA copy numbers in cancer samples in patients with chronic lymphocytic leukemia, lung squamous cell carcinoma and pancreatic adenocarcinoma, but decreased copy numbers in cancer samples in patients with kidney clear cell carcinoma, hepatocellular carcinoma and myeloproliferative neoplasm (Fig. 5d). At face value, the distinct patterns in different cancer types may be due to cancer-specific oncogenic stimulation, metabolic activity and mitochondrial malfunctions. For example, a recent study12 suggested that significantly decreased mtDNA copy number in kidney clear cell cancer may be due to downregulation of peroxisome proliferator-activated receptor-γ coactivator 1α (a central regulator of mitochondrial biogenesis) by hyperactivated hypoxia-inducible factor 1α, which is most frequently mutated and activated in this disease38. However, since the available mtDNA copy numbers in normal tissues are average values from mixtures of many heterogeneous cell types with unknown relative contributions, a direct comparison between tumor and adjacent normal tissues should be interpreted cautiously.
a, Distributions of mtDNA copy number by cancer tissue type. Sample numbers with mtDNA copy-number information available are labeled on the top, with the median mtDNA copy numbers marked as red bars. b, Distinct mtDNA copy-number distributions for cancer types derived from the kidney (top) and brain (bottom). n = number of samples with mtDNA copy number. P values were generated by ANOVA. c, mtDNA copy numbers with and without truncating mutations in mtDNA genes. n = number of samples with both mtDNA copy number and somatic mutation information available. P values were based on ANOVA, adjusting for cancer types. d, Paired copy-number comparison of tumor and matching normal tissue samples. n = number of matching normal tissue and cancer sample pairs. Raw P values were determined by two-sided Wilcoxon signed-rank test, then adjusted for FDR (**FDR < 0.01; ***FDR < 0.001). e, Correlation of mtDNA copy number with patient age in prostate cancer. n = number of samples with cancer mtDNA copy number and patient age information available. Correlations and P values are based on Spearman’s rank correlation. f, Correlation of mtDNA copy number with cancer stage in chronic lymphocytic leukemia. n = number of samples with mtDNA copy number and stage information. g, Focal copy gain and loss caused by structural variations in three cancer samples. Green lines represent focal loss, whereas red lines represent tandem duplication. In all boxplots, the boundaries of the boxes mark the first and third quartile, with the median in the center, and whiskers extending to 1.5× the interquartile range from the boundaries.
To assess the potential biomedical significance of mtDNA copy numbers, we examined their correlations with key clinical variables. We found significant positive correlations between the mtDNA copy number and the patient’s age at diagnosis in prostate (Spearman’s rank, Rs = 0.31; P < 1.7 × 10−4; Fig. 5e), colorectal and skin cancers (Supplementary Fig. 17). In contrast, we observed negative correlations of normal blood mtDNA copy number with patient age in most cases (Supplementary Fig. 18). We further observed correlations between mtDNA copy number and tumor stage in multiple cancer types (Fig. 5f and Supplementary Fig. 19).
Using WGS data, we examined the focal copy gain and loss in the mitochondrial genomes that were known to be present in prostate cancers and aged tissues39. Of the 2,658 cancer samples, three (0.11%) showed notable structural variants in the mtDNA (Fig. 5g). For example, a pancreatic cancer case (sample ID: SP76017) harbored a ~3.4-kb-long mtDNA loss that truncated ribosomal RNA and ND1 genes. The VAF of this mutant mtDNA was estimated at 63%. Similarly, a melanoma case (sample ID: SP127680) showed tandem duplication of an mtDNA segment of ~4 kb, with 100% VAF. Thus, our analysis identified structural variants in mtDNA genomes based on WGS.
Co-expression network analysis of mitochondrial genes
To understand the functional impact of 13 mtDNA genes in cancers, we quantified the gene expression levels using RNA-seq data profiled from 4,689 TCGA tumor samples of 13 cancer types (Supplementary Table 4). The correlation between the gene expression levels and the mtDNA copy number varied by cancer type (Supplementary Fig. 20). Among the cancer types, the mtDNA genes were highly expressed in the three types of kidney cancer (chromophobe, papillary and clear cell) but weakly expressed in the three types of squamous cell carcinoma (cervical, lung and head and neck) (Fig. 6a). This observation was partially due to the relative abundance of mtDNA copy number across cancer types and is consistent with a study of normal tissues40.
a, Left: heat map of the expression levels of 13 mtDNA genes of 13 cancer types. Right: bar plot showing the sample sizes for each cancer type. b, Commonly enriched pathways identified by co-expression with mtDNA genes in different cancer types. Borders of cells with FDR < 0.05 are highlighted in yellow. c, mtDNA gene-centric pan-cancer co-expression network. The pie chart colors at each node indicate occurrence of the node in cancer types of the corresponding colors. Green borders, nuclear genes; blue borders, mitochondrial genes. Node size is proportional to the number of direct neighbors (connectivity) of the node. Thickness of the edge is proportional to the frequency of this edge being observed across all cancer types. Edges are colored according to the connection type (gray: mtDNA gene–mtDNA gene connection; magenta: mtDNA gene–nuclear gene connection). BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, brain lower-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; THCA, thyroid carcinoma.
To gain more insight into the functions of mtDNA genes and their related nuclear genes and pathways, for each cancer type, we used the weighted gene co-expression network analysis (WGCNA) package41 to build a weighted gene co-expression network that consisted of both nuclear genes and mitochondrial genes (Methods). We then performed gene set enrichment analysis (GSEA)42 based on the rank of all nuclear genes by measuring their edge strength to a mitochondrial gene in the co-expression network. We found oxidative phosphorylation to be the top-ranked enriched pathway, and to be enriched in eight out of the 13 cancer types examined (FDR < 0.05), highlighting the essential role of mitochondrial genes in energy generation (Fig. 6b). Pathways related to the cell cycle (MYC targets, mitotic spindle, G2/M checkpoint and E2F targets) and DNA repair were also enriched in multiple cancer types (Fig. 6b), consistent with the established notion that mtDNA plays an important role in these pathways37,43.
We also examined the mtDNA-centric co-expression networks (Fig. 6c and Methods). Across cancer types, the mtDNA genes were almost always strongly interconnected, which is expected since they are transcribed as long polycistronic precursor transcripts44. Interestingly, several clinically actionable genes were among the neighboring genes that showed strong co-expression patterns with mtDNA genes (Fig. 6c and Supplementary Fig. 21). For example, AR, EGFR, DDR2 and MAP2K2 were connected with mtDNA genes in prostate cancer, and TMPRSS2, NF1, PIK3CA, BRCA1 and TOP1 were the top neighbors of mtDNA genes in multiple cancer types. This correlation-based analysis does not necessarily suggest causality, and further efforts are needed to investigate these relationships. Elucidating the underlying mechanisms may lay a foundation for developing mtDNA-related cancer therapy.
An open-access Cancer Mitochondrial Atlas data portal
To facilitate mitochondria-related biological discoveries and clinical applications, we developed an open-access, user-friendly data portal, The Cancer Mitochondrial Atlas (TCMA), for fluent exploration of the various types of molecular data characterized in this study (Supplementary Fig. 22). The data portal can be accessed at http://bioinformatics.mdanderson.org/main/TCMA:Overview. There are four modules in TCMA: somatic mutations, nuclear transfer, copy number and gene expression. The first three modules are based on the ICGC WGS data and provide detailed annotations for the corresponding features of each cancer sample. The last module is based on TCGA RNA-seq data and provides an interactive interface through which users can visualize the co-expression network with convenient navigation and zoom features. Not only can users browse and query the molecular data by cancer type, they can also download all of the data for their own analysis.
Discussion
This work characterizes the cancer mitochondrial genome in a comprehensive manner, including somatic mutations, nuclear transfer, copy number, structural variants and mtDNA gene expression. Because of the ultra-high coverage of mtDNA from the WGS data and the large number of patient samples surveyed, our study provides a definitive landscape of mtDNA somatic mutations and identifies several unique features. First, we report hypermutated mitochondrial cases, highlighting the dynamic mutational processes in this tiny genome. Second, our systemic analysis of mitochondrial genomes has firmly shown that several cancer types are enriched for high-allele-frequency truncating mutations, including previously reported kidney chromophobe30,45 as well as newly identified kidney papillary, and thyroid and colorectal cancers. Interestingly, the thyroid and kidney are the most frequent sites of oncocytomas, which are rare, benign tumors characterized by frequent nuclear chromosomal aneuploidy as well as vast accumulation of defective mitochondria45,46, further assuring the functional association between mitochondrial inactivation and the pathogenesis of these cancer types. Third, in contrast with the diversified mutational signatures observed in the nuclear genomes of different cancers20, mtDNAs show very similar mutational signatures regardless of cancer tissue origins: predominantly G>A and T>C substitutions on the L strand. This monotonous pattern may partially stem from different mutational generators and DNA repair processes between the nucleus and mitochondria9,47,48. Due to their large numbers of copies per cell, mitochondria may simply remove mtDNA damaged from external mutagens (for example, ultraviolet radiation, tobacco smoking and reactive oxygen species) through autophagy and other mitochondrial dynamic mechanisms49, rather than employing a complex array of repair proteins as in the nucleus.
One unique aspect of our study is the integrative analysis of mitochondrial molecular alterations with those in the nuclear genome that are characterized by the PCAWG Consortium. We found that: (1) high-allele-frequency truncating mtDNA mutations are mutually exclusive to mutated cancer genes in kidney cancer; (2) mtDNA nuclear transfers are associated with increased numbers of structural variants in the nuclear genome; and (3) mtDNA co-expressed nuclear genes are enriched in several processes critical for tumor development. These results indicate that the mitochondrial genome is an essential component in understanding the complex molecular patterns observed in cancer genomes and helping to pinpoint potential cancer driver events. Our results, such as the nuclear transfer of mtDNA into a therapeutic target gene, correlations of mtDNA copy numbers with clinical variables, and the co-expression of mtDNA and clinically actionable genes, underscore the clinical importance of mitochondria.
Taken together, this study has untangled and characterized the full spectrum of molecular alterations of mitochondria in human cancers. Our analyses have provided essentially complete catalogs of somatic mtDNA alterations in cancers, including substitutions, indels, copy-number alterations and structural variants. Furthermore, we have developed a user-friendly web resource to enable the broader biomedical community to capitalize on our results. These efforts lay a foundation for translating mitochondrial biology into clinical investigations.
Methods
Data generation and collection
We extracted BAM files of mtDNA sequencing reads from the whole-genome alignment files of 2,658 cancer samples and their matched normal tissue samples generated by the PCAWG Consortium. BWA was used to align the reads to the human reference genome (hs37d5). From the CGHub, we obtained TCGA RNA-seq BAM files of 13 cancer types, all of which employed paired-end sequencing strategies. We used Cufflinks to quantify the messenger RNA expression levels (in fragments per kilobase per million mapped fragments) of the 13 mitochondrial protein-coding genes. We obtained the nuclear somatic mutations and annotated driver mutations of corresponding samples as described17.
Somatic mutation calling
The nuclear genome mutations were called using the Sanger pipeline, provided by the PCAWG. The mitochondrial variants were initially called using VarScan2 (ref. 50) and the same parameter setting as previously reported9: --strand-filter 1 (mismatches should be reported by both forward and reverse reads), --min-var-freq 0.01 (minimum VAF 1%), --min-avg-qual 20 (minimum base quality 20), --min-coverage × and --min-reads2 ×). We applied a series of downstream bioinformatic filters to further remove false positives as follows (Supplementary Fig. 2a).
First, we filtered germline polymorphisms and false positive calls (for example, frequent mapping errors due to known mtDNA homopolymers, candidates with substantial mapping strand bias and candidates with substantial mutant alleles in the matched normal sample). For analytic simplicity, we removed multi-allelic mtDNA mutations and back mutations from the non-reference to the reference allele. After this filtration step, we obtained 10,083 somatic substitution candidates.
Second, we examined DNA cross-contamination because even minor DNA cross-contamination (that is, contamination level < 3%) would generate many low-VAF false positive calls that are in fact germline polymorphisms from the contaminating sample. We tested whether mtDNA somatic mutations detected from a cancer sample show greater overlap with known mtDNA polymorphisms than expected from the overall average rate (73.5%; 3,922/5,337 substitutions) using the binomial test with a cutoff P < 0.01. From this step, we removed 96 samples with evidence of DNA cross-contamination (harboring 935 known mutations out of 1,131 known mutation candidates).
Third, we examined the overall mtDNA substitution signatures in the 96 possible mutation classes. We removed four samples with extremely high proportions of C>G substitutions with strong sequence context bias (at CpCpN>CpApN; most frequently at CpCpG>CpApG; Supplementary Fig. 2b). This spectrum is known to arise from artificial guanine oxidation during sequencing library preparation steps19 with low VAF (1–2%). We explicitly removed these samples from further analyses.
Then, we examined the possibility of false positive calls due to mismapping of reads from inherited nuclear mtDNA-like sequences (known as numts) not represented in the human reference genome18, especially when the specific numts regions were amplified in the cancer nuclear genome. These mutation candidates showed some specific features: (1) they appeared as highly recurrent mtDNA somatic mutations among multiple samples; (2) VAFs in mitochondria were only slightly higher than our 1% cutoff criteria; and (3) the matched normal samples also had small but substantial numbers of mutation allele counts. To remove these false positive calls, we applied two statistical tests of: (1) whether the VAF of a mutation candidate in the matched normal sequences was within the normal range (<0.0024; the cutoff is determined by the median VAF of all mutation candidates +2× the interquartile range); and (2) whether:
was within the normal range (<0.0357; the cutoff is determined by the median VAF of all mutation candidates +2× the interquartile range), where Nmut is the mutation allele count, RD is the average read depth for the nuclear genome, and nor and tum are normal and matched tumor tissues, respectively. When a mutation appeared to be an outlier according to both criteria, we removed the candidate from our downstream analyses.
In our previous study9, we could not detect mutations under a 3% VAF cutoff because mtDNA was sequenced with a read depth of ~100× from the majority of samples surveyed. Taking advantage of the ultra-high depth (>8,000×) in this study, we used a 1% VAF cutoff to obtain better sensitivity. We found 2,133 more substitutions when the VAF was between 1 and 3%. Because of the ultra-high depth, even 1% VAF mutations were considered to be specific, and were supported by a high number (n = ~80) of mutation alleles. We confirmed the high specificity of these mutations using the unique mtDNA mutational signatures robustly observed even from these low-VAF mutations: (1) the mutational spectrum is generally consistent with those from higher heteroplasmic levels of mutations (that is, VAFs from 3–10% and 10–100%); (2) we observed the absolute dominance of C>T and T>C substitutions in the expected trinucleotide contexts (NpCpG for C>T and NpTpC for T>C substitutions); and (3) we also observed extreme replication strand bias (Supplementary Fig. 3). These features would not be observed if contaminations resulted in many false positive calls. To assess the factors affecting the mutation frequency of the 13 coding genes, we performed the sample-level analysis using log-linear modeling: we assigned the binary mutation indicator (1: with mutation; 0: without mutation) to each sample for each gene and then fit this binary response variable to a logistic regression model, including cancer type, gene identity and their interaction as explanatory variables, which were later summarized using ANOVA. In addition, within each cancer type, we used Spearman’s rank correlation to assess the association between the numbers of nuclear and mtDNA somatic mutations, as well as their individual association with patient age.
Truncating mutation analysis
Taking into account the mtDNA-specific mutational signature, we examined the dN/dS ratio for mtDNA missense substitutions as reported previously9. We defined truncating mutations as those that lead to truncated protein products (that is, nonsense mutations and frameshift indels), and accordingly categorized the samples into the truncating group (bearing at least one truncating mutation with VAF ≥ 60%). The ND5 protein domain information was obtained from Pfam (http://pfam.xfam.org/protein/P03915). The cancer gene census list was obtained from http://cancer.sanger.ac.uk/cosmic/download. Cancer census genes with recurrent somatic mutations in kidney chromophobe and kidney papillary cancers were selected for analysis of mutual exclusivity and heat-map representation. One sample with a nuclear DNA hypermutator phenotype was excluded from this analysis. To examine the functional consequences of mtDNA truncating mutations, we performed GSEA based on the ranks of differentially expressed genes between samples with and samples without mtDNA truncating mutations for kidney chromophobe, kidney papillary, colorectal and thyroid cancers and their combination, and identified significantly enriched pathways at FDR = 0.05.
SMNT analysis
We examined the WGS data from the cancer and matched control tissue samples using a pipeline for the identification of mtDNA translocation to the nuclear genome, as reported previously11. The specificity was shown to be 100% in the previous study11. Briefly, we extracted and clustered discordant reads from cancer genomes, where one end aligned to nuclear DNA and the other aligned to mtDNA. Then, to determine the nucleotide resolution breakpoints, we searched for split reads near putative breakpoint junctions (1,000 base pairs upstream and downstream), where a fraction of a single read aligned to genomic DNA near the junctions and the rest aligned to mtDNA. All filtering criteria were the same as previously reported, except that we did not use BLAT51 for split-read detection because the BWA-MEM alignment tool used to map all pan-cancer samples fundamentally enables split-read mapping. We removed candidate mitochondrion–nuclear DNA junctions that overlapped with clusters from matched and unmatched normal samples and/or known human SMNTs—a combined set from the human reference genome (hg19; n = 123) and a published study52 (n = 766)—because the source of the mtDNA sequence fused to the nuclear genome might be SMNTs rather than real mitochondria in the cytoplasm of cells. We obtained the PCAWG Structural Variation Working Group16 and compared the samples with and without SMNTs by t-test. To study the relationship of SMNTs and structural variant breakpoints, we randomly chose the same number of structural variant breakpoints from each sample 100 times to estimate the random expectation.
MtDNA copy-number analysis
To better estimate the mtDNA copy number for cancer samples, we employed the following formula, which incorporates both tumor purity and ploidy information:
where f is the tumor purity (ranging from 0 to 1, where 1 stands for pure cancer cells and 0 stands for pure normal cells), CN is the mtDNA copy number, coverage_depthmtDNA and coverage_depthgDNA are the mean coverage depths for mtDNA and the nuclear genome in individual WGS BAM files, respectively, and ploidycancer is the number of sets of chromosomes in tumor cells, while ploidy in the normal cells is 2. Both f and ploidycancer were obtained using allele-specific copy-number analysis of tumors estimation53, provided by the PCAWG Consortium. Donors with multiple samples were preselected so that each donor came with one representative primary cancer sample. We excluded cancer samples with low purity (<0.4, estimated by allele-specific copy-number analysis of tumors) for further downstream analyses. We used ANOVA (if there were more than two cancer types) or t-test to compare the mtDNA copy number of cancer types derived from the same tissue. Since many of the normal samples were from blood, we focused on the cancer types with at least ten samples from the normal tissue adjacent to the tumor in order to compare the mtDNA copy number of the paired cancer and normal samples. We used the Wilcoxon signed-rank test to compare the mtDNA copy number for each selected cancer type and further adjusted the raw P values based on the FDR. To assess the correlation of mtDNA copy number with truncating mutations, we employed ANOVA (with the cancer type included in the model, to account for its potential effect). We assessed the correlations of the mtDNA copy number with the patient’s age, overall survival time and cancer stage using Spearman’s rank correlation, Cox model/log-rank test and ANOVA, respectively. We log2-transformed the mtDNA copy-number values when using ANOVA and the t-test, to conform to the normality assumption.
mtDNA structural variation analysis
To investigate large deletions or duplications in the mtDNA genome, we sought the read-depth change of tumor mtDNA sequences using normal mtDNA sequences as a reference. To this end, we calculated the normalized depth of mtDNA loci in 100-base pair-sized bins from all of the normal samples. Then, we calculated the deviation of mtDNA read depth in each tumor sample. When ten bins were consecutively increased or decreased in the relative depth sufficiently (z score > 3), we considered the region as a structural variation candidate. From all of the candidates, we sought discordant paired-end reads, or breakpoint-spanning reads, which strongly support structural variations11.
Co-expression analysis
For each cancer type, we used the WGCNA package41 to build a weighted gene co-expression network that contains ~20,000 nodes (including both nuclear genes and mitochondrial genes). The key parameter, β, for a weighted network construction was optimized to maintain both the scale-free topology and sufficient node connectivity, as recommended in the manual. In such a network, any two genes were connected and the edge weight was determined by the topology overlap measure provided in WGCNA. This measure considered not only the expression correlation between two partner genes, but also how many ‘friends’ the two genes shared. The weights ranged from 0 to 1, which reflected the strength of the connection between the two genes. To identify mitochondria-related pathways, we performed GSEA42 on the basis of the full set of nuclear protein-coding genes, ranked on the basis of the weights of the edge connecting the mitochondrial genes, and detected significant pathways at FDR = 0.05. To construct the mitochondria-centric network, we focused on the top 500 neighboring genes that showed the strongest connections with the mitochondrial genes, with a minimum weight of 0.05. Among these neighboring genes, we detected the clinically actionable genes (defined as FDA-approved therapeutic targets and their relevant predictive markers54) in at least one of the cancer types we surveyed. We examined the correlations of mtDNA gene expression levels with mtDNA copy numbers using Spearman’s rank correlations.
TCMA data portal construction
We stored the precalculated mtDNA molecular data (including mtDNA mutation, nuclear transfer, copy number and expression) in a database of CouchDB. The Web interface was implemented by JavaScript, tables were visualized by DataTables, and the co-expression network visualization was implemented by Cytoscape Web.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Somatic and germline variant calls, mutational signatures, subclonal reconstructions, transcript abundance, splice calls and other core data generated by the ICGC/TCGA PCAWG Consortium are described here14 and are available for download at https://dcc.icgc.org/releases/PCAWG. Additional information on accessing the data, including raw read files, can be found at https://docs.icgc.org/pcawg/data/. In accordance with the data access policies of the ICGC and TCGA projects, most molecular, clinical and specimen data are in an open tier that does not require access approval. To access information with the potential to identify individuals, such as germline alleles and underlying sequencing data, researchers will need to apply to the TCGA Data Access Committee via the Database of Genotypes and Phenotypes (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login) for access to the TCGA portion of the dataset, and to the ICGC Data Access Compliance Office (http://icgc.org/daco) for the ICGC portion. In addition, to access somatic single-nucleotide variants derived from TCGA donors, researchers will also need to obtain Database of Genotypes and Phenotypes authorization. Derived datasets described specifically in this study are available from the TCMA data portal (http://bioinformatics.mdanderson.org/main/TCMA:Overview).
Code availability
The core computational pipelines used by the PCAWG Consortium for alignment, quality control and variant calling are available to the public at https://dockstore.org/search?search=pcawg under GNU General Public License version 3.0, which allows for reuse and distribution.
Change history
11 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41588-020-0587-4
27 April 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41588-020-0629-y
21 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41588-023-01317-x
11 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41588-020-0587-4
27 April 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41588-020-0629-y
References
Schon, E. A., DiMauro, S. & Hirano, M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878–890 (2012).
Smeitink, J., van den Heuvel, L. & DiMauro, S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2, 342–352 (2001).
Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981).
Brandon, M., Baldi, P. & Wallace, D. C. Mitochondrial mutations in cancer. Oncogene 25, 4647–4662 (2006).
Zong, W. X., Rabinowitz, J. D. & White, E. Mitochondria and cancer. Mol. Cell 61, 667–676 (2016).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hengartner, M. O. The biochemistry of apoptosis. Nature 407, 770–776 (2000).
Larman, T. C. et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl Acad. Sci. USA 109, 14087–14091 (2012).
Ju, Y. S. et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 3, e02935 (2014).
Stewart, J. B. et al. Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 11, e1005333 (2015).
Ju, Y. S. et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res. 25, 814–824 (2015).
Reznik, E. et al. Mitochondrial DNA copy number variation across human cancers. eLife 5, e10769 (2016).
Hopkins, J. F. et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 8, 656 (2017).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature https://doi.org/10.1038/s41586-020-1969-6 (2020).
The Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Li, Y. et al. Patterns of somatic structural variation in human cancer genomes. Nature https://doi.org/10.1038/s41586-019-1913-9 (2020).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,693 cancer whole genomes. Nature https://doi.org/10.1038/s41586-020-1965-x (2020).
Dayama, G., Emery, S. B., Kidd, J. M. & Mills, R. E. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 42, 12640–12649 (2014).
Costello, M. et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67 (2013).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Kennedy, S. R., Salk, J. J., Schmitt, M. W. & Loeb, L. A. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9, e1003794 (2013).
Tanaka, M. & Ozawa, T. Strand asymmetry in human mitochondrial DNA mutations. Genomics 22, 327–335 (1994).
Zheng, W., Khrapko, K., Coller, H. A., Thilly, W. G. & Copeland, W. C. Origins of human mitochondrial point mutations as DNA polymerase γ-mediated errors. Mutat. Res. 599, 11–20 (2006).
Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015).
Coller, H. A. et al. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat. Genet. 28, 147–150 (2001).
The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
The Cancer Genome Atlas Research Network et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Pak, J. W., Vang, F., Johnson, C., McKenzie, D. & Aiken, J. M. MtDNA point mutations are associated with deletion mutations in aged rat. Exp. Gerontol. 40, 209–218 (2005).
Davis, C. F. et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 (2014).
Caro, P. et al. Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion 10, 479–486 (2010).
Chen, D., Xue, W. & Xiang, J. The intra-nucleus integration of mitochondrial DNA (mtDNA) in cervical mucosa cells and its relation with c-myc expression. J. Exp. Clin. Cancer Res. 27, 36 (2008).
Srinivasainagendra, V. et al. Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma. Genome Med. 9, 31 (2017).
Cui, H. et al. Association of decreased mitochondrial DNA content with the progression of colorectal cancer. BMC Cancer 13, 110 (2013).
Dickinson, A. et al. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 20, 1644–1653 (2013).
Van Osch, F. H. et al. Mitochondrial DNA copy number in colorectal cancer: between tissue comparisons, clinicopathological characteristics and survival. Carcinogenesis 36, 1502–1510 (2015).
McBride, H. M., Neuspiel, M. & Wasiak, S. Mitochondria: more than just a powerhouse. Curr. Biol. 16, R551–R560 (2006).
Vyas, S., Zaganjor, E. & Haigis, M. C. Mitochondria and cancer. Cell 166, 555–566 (2016).
Lott, M. T. et al. mtDNA variation and analysis using mitomap and mitomaster. Curr. Protoc. Bioinformatics 44, 1.23.1–1.23.26 (2013).
Mercer, T. R. et al. The human mitochondrial transcriptome. Cell 146, 645–658 (2011).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Koczor, C. A. et al. Mitochondrial DNA damage initiates a cell cycle arrest by a Chk2-associated mechanism in mammalian cells. J. Biol. Chem. 284, 36191–36201 (2009).
Ojala, D., Montoya, J. & Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 (1981).
Joshi, S. et al. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 13, 1895–1908 (2015).
Gasparre, G., Romeo, G., Rugolo, M. & Porcelli, A. M. Learning from oncocytic tumors: why choose inefficient mitochondria? Biochim. Biophys. Acta 1807, 633–642 (2011).
Clayton, D. A., Doda, J. N. & Friedberg, E. C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA 71, 2777–2781 (1974).
Haradhvala, N. J. et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549 (2016).
Bess, A. S., Crocker, T. L., Ryde, I. T. & Meyer, J. N. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 40, 7916–7931 (2012).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Simone, D., Calabrese, F. M., Lang, M., Gasparre, G. & Attimonelli, M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics 12, 517 (2011).
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc. Natl Acad. Sci. USA 107, 16910–16915 (2010).
Van Allen, E. M. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 20, 682–688 (2014).
Acknowledgements
This study was partially supported by an MD Anderson Cancer Center Faculty Scholar Award (to H.L.), the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine (to J.N.W.), an Institute for Information and Communications Technology Promotion grant funded by the Korean government (Ministry of Science, ICT and Future Planning) (B0101-15-0104; the development of a supercomputing system for genome analysis), the Korea Health Technology Research and Development Project (through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea) (HI14C0072 to H.-L.K. and HI17C1836 to Y.S.J.) and the Korean National Research Foundation (NRF-2016R1D1A1B03934110 and NRF-2017R1A2B2012796). We also thank the Electronics and Telecommunications Research Institute in Korea for its commitment to the ICGC PCAWG projects, the MD Anderson Cancer Center High-Performance Computing Core Facility for computing, and L. Chastain for editorial assistance. We acknowledge the contributions of the members of the many clinical networks across ICGC and TCGA who provided samples and data to the PCAWG Consortium, and the contributions of the Technical Working Group and the Germline Working Group of the PCAWG Consortium for collation, realignment and harmonized variant calling of the cancer genomes used in this study. We thank the patients and their families for participation in the individual ICGC and TCGA projects.
Author information
Authors and Affiliations
Consortia
Contributions
K.P., P.J.C. and H.L. conceived and designed the project. Y. Yuan, Y.S.J., Y.K., J.L., Y.W., C.J.Y., Y. Yang, I.M., C.J.C., J.N.W., Y.X., L.H., H.-L.K., H.N., K.P., P.J.C. and H.L. contributed to the data analysis. Y. Yuan, Y.S.J., Y.K. and H.L. wrote the manuscript with input from all other authors. H.L. supervised the entire project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–22 and Tables 1–4
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, Y., Ju, Y.S., Kim, Y. et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet 52, 342–352 (2020). https://doi.org/10.1038/s41588-019-0557-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0557-x
This article is cited by
-
Leveraging new methods for comprehensive characterization of mitochondrial DNA in esophageal squamous cell carcinoma
Genome Medicine (2024)
-
Mitochondrial DNA mutations drive aerobic glycolysis to enhance checkpoint blockade response in melanoma
Nature Cancer (2024)
-
Mitochondrial DNA mutations in Medulloblastoma
Acta Neuropathologica Communications (2023)
-
A systematic comparison of human mitochondrial genome assembly tools
BMC Bioinformatics (2023)
-
Next-generation sequencing reveals mitogenome diversity in plasma extracellular vesicles from colorectal cancer patients
BMC Cancer (2023)