Abstract
Metastasis is the primary cause of cancer mortality, and cancer frequently metastasizes to the liver. It is not clear whether liver immune tolerance mechanisms contribute to cancer outcomes. We report that liver metastases diminish immunotherapy efficacy systemically in patients and preclinical models. Patients with liver metastases derive limited benefit from immunotherapy independent of other established biomarkers of response. In multiple mouse models, we show that liver metastases siphon activated CD8+ T cells from systemic circulation. Within the liver, activated antigen-specific Fas+CD8+ T cells undergo apoptosis following their interaction with FasL+CD11b+F4/80+ monocyte-derived macrophages. Consequently, liver metastases create a systemic immune desert in preclinical models. Similarly, patients with liver metastases have reduced peripheral T cell numbers and diminished tumoral T cell diversity and function. In preclinical models, liver-directed radiotherapy eliminates immunosuppressive hepatic macrophages, increases hepatic T cell survival and reduces hepatic siphoning of T cells. Thus, liver metastases co-opt host peripheral tolerance mechanisms to cause acquired immunotherapy resistance through CD8+ T cell deletion, and the combination of liver-directed radiotherapy and immunotherapy could promote systemic antitumor immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw data for single-cell sequencing are deposited at the NCBI Gene Expression Omnibus (GSE157600). Further information and requests for resources and reagents should be directed to the corresponding authors. All requests for raw and analyzed data and materials will be promptly reviewed by the corresponding authors to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement. Patient-related data not included in the paper may be subject to patient confidentiality. Source data have been provided with this paper for Figs. 1–6 as well as Extended Figs. 1–9.
References
Mehlen, P. & Puisieux, A. Metastasis: a question of life or death. Nat. Rev. Cancer 6, 449–458 (2006).
Disibio, G. & French, S. W. Metastatic patterns of cancers: results from a large autopsy study. Arch. Pathol. Lab. Med. 132, 931–939 (2008).
Li, F. & Tian, Z. The liver works as a school to educate regulatory immune cells. Cell. Mol. Immunol. 10, 292–302 (2013).
Doherty, D. G. Immunity, tolerance and autoimmunity in the liver: a comprehensive review. J. Autoimmun. 66, 60–75 (2016).
Crispe, I. N. Hepatic T cells and liver tolerance. Nat. Rev. Immunol. 3, 51–62 (2003).
Crispe, I. N., Dao, T., Klugewitz, K., Mehal, W. Z. & Metz, D. P. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunol. Rev. 174, 47–62 (2000).
Zou, W., Wolchok, J. D. & Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv4 (2016).
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Lin, H. et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade–mediated tumor regression. J. Clin. Invest. 128, 805–815 (2018).
Wei, S. C., Duffy, C. R. & Allison, J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018).
Curiel, T. J., Wei, S., Dong, H., Alvarez, X. & Cheng, P. Blockade of B7-H1 improves myeloid dendritic cell–mediated antitumor immunity. Nat. Med. 9, 562–567 (2003).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Jiao, S. et al. Differences in tumor microenvironment dictate T helper lineage polarization and response to immune checkpoint therapy. Cell 179, e13 (2019).
Spranger, S. & Gajewski, T. F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 18, 139–147 (2018).
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer–immune set point. Nature 541, 321 (2017).
Formenti, S. C. et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat. Med. 24, 1845–1851 (2018).
Theelen, W. S. et al. Effect of pembrolizumab after stereotactic body radiotherapy vs pembrolizumab alone on tumor response in patients with advanced non–small cell lung cancer: results of the PEMBRO-RT phase 2 randomized clinical trial. JAMA Oncol. 5, 1276–1282 (2019).
Golden, E. B., Demaria, S., Schiff, P. B., Chachoua, A. & Formenti, S. C. An abscopal response to radiation and ipilimumab in a patient with metastatic non-small cell lung cancer. Cancer Immunol. Res 1, 365–372 (2013).
Twyman-Saint Victor, C. et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377 (2015).
Vanpouille-Box, C. et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017).
Deng, L. et al. Irradiation and anti–PD-L1 treatment synergistically promote antitumor immunity in mice. J. Clin. Invest. 124, 687–695 (2014).
Huang, A. C. et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017).
Yarchoan, M. et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight 4, e126908 (2019).
Petrelli, F. et al. Prognostic and predictive role of elevated lactate dehydrogenase in patients with melanoma treated with immunotherapy and BRAF inhibitors: a systematic review and meta-analysis. Melanoma Res. 29, 1 (2019).
Tang, H. et al. PD-L1 on host cells is essential for PD-L1 blockade–mediated tumor regression. J. Clin. Invest. 128, 580–588 (2018).
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013).
John, B. & Crispe, I. N. Passive and active mechanisms trap activated CD8+ T cells in the liver. J. Immunol. 172, 5222–5229 (2004).
Topalian, S. L. et al. Five-year survival and correlates among patients with advanced melanoma, renal cell carcinoma, or non–small cell lung cancer treated with nivolumab. JAMA Oncol. 5, 1411–1420 (2019).
Robinson, D. R. et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Medina-Echeverz, J., Eggert, T., Han, M. & Greten, T. F. Hepatic myeloid-derived suppressor cells in cancer. Cancer Immunol. Immunother. 64, 931–940 (2015).
Li, W. et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB Isoform in triple-negative breast cancer. Cell Metab. 28, e6 (2018).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, e29 (2020).
Saio, M., Radoja, S., Marino, M. & Frey, A. B. Tumor-infiltrating macrophages induce apoptosis in activated CD8+ T cells by a mechanism requiring cell contact and mediated by both the cell-associated form of TNF and nitric oxide. J. Immunol. 167, 5583–5593 (2001).
Zhu, J., Petit, P.-F. & Van den Eynde, B. J. Apoptosis of tumor-infiltrating T lymphocytes: a new immune checkpoint mechanism. Cancer Immunol. Immunother. 68, 835–847 (2019).
Blériot, C. & Ginhoux, F. Understanding the heterogeneity of resident liver macrophages. Front. Immunol. 10, 2694 (2019).
David, B. A. et al. Combination of mass cytometry and imaging analysis reveals origin, location, and functional repopulation of liver myeloid cells in mice. Gastroenterology 151, 1176–1191 (2016).
Jackson, W. C. et al. Comparison of stereotactic body radiation therapy and radiofrequency ablation in the treatment of intrahepatic metastases. Int. J. Radiat. Oncol.*Biol.*Phys. 100, 950–958 (2018).
Peng, D. et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015).
Tumeh, P. C. et al. Liver metastasis and treatment outcome with anti-PD-1 monoclonal antibody in patients with melanoma and NSCLC. Cancer Immunol. Res 5, 417–424 (2017).
Li, H. et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 56, 1342–1351 (2012).
Zeng, Z. et al. Interferon-γ facilitates hepatic antiviral T cell retention for the maintenance of liver-induced systemic tolerance. J. Exp. Med. 213, 1079–1093 (2016).
Limmer, A. et al. Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat. Med. 6, 1348–1354 (2000).
Warren, A. et al. T lymphocytes interact with hepatocytes through fenestrations in murine liver sinusoidal endothelial cells. Hepatology 44, 1182–1190 (2006).
Goubier, A. et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity 29, 464–475 (2008).
Ma, C. et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Horst, A. K., Neumann, K., Diehl, L. & Tiegs, G. Modulation of liver tolerance by conventional and nonconventional antigen-presenting cells and regulatory immune cells. Cell. Mol. Immunol. 13, 277–292 (2016).
Lee, J. C. et al. Regulatory T cell control of systemic immunity and immunotherapy response in liver metastasis. Sci. Immunol. 5, eaba0759 (2020).
Reck, M. et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir. Med 7, 387–401 (2019).
Lang, X. et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Disco. 9, 1673–1685 (2019).
Welsh, J. W. et al. Phase II trial of Ipilimumab with stereotactic radiation therapy for metastatic disease: outcomes, toxicities, and low-dose radiation-related abscopal responses. Cancer Immunol. Res. 7, 1903–1909 (2019).
Hosein, A. N., Brekken, R. A. & Maitra, A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 17, 487–505 (2020).
Mayo, C. S. et al. The big data effort in radiation oncology: Data mining or data farming? Adv. Radiat. Oncol. 1, 260–271 (2016).
Zhang, Q. et al. Inhibition of ATM increases interferon signaling and sensitizes pancreatic cancer to immune checkpoint blockade therapy. Cancer Res. 79, 3940–3951 (2019).
Sijts, A. J., Ossendorp, F. & Mengede, E. A. Immunodominant mink cell focus-inducing murine leukemia virus (MuLV)-encoded CTL epitope, identified by its MHC class I-binding motif, explains MuLV specificity of MCF-directed cytotoxic T lymphocytes. J. Immunol. 152, 106–116 (1994).
Yu, J. et al. The differential organogenesis and functionality of two liver-draining lymph nodes in mice. J. Autoimmun. 84, 109–121 (2017).
Zheng, M., Yu, J. & Tian, Z. Characterization of the liver-draining lymph nodes in mice and their role in mounting regional immunity to HBV. Cell. Mol. Immunol. 10, 143–150 (2013).
Billi, A. C. et al. The female-biased factor VGLL3 drives cutaneous and systemic autoimmunity. JCI Insight 4, e127291 (2019).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Mould, K. J., Jackson, N. D., Henson, P. M., Seibold, M. & Janssen, W. J. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 4, e126556 (2019).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Dwyer, D. F., Barrett, N. A. & Austen, K. F., Immunological Genome Project Consortium. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17, 878–887 (2016).
Peng, H. et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123, 1444–1456 (2013).
Imai, K. & Ratkovic, M. Covariate balancing propensity score. J. R. Stat. Soc. Ser. B Stat. Methodol. 76, 243–263 (2014).
Acknowledgements
This work was supported in part by research grants from the NIH/NCI grants for WZ (CA248430, CA123088, CA099985, CA193136, and CA152470); AC (1UM1HG006508); TSL (U01CA216449); IEN (CA233487); FW (S10OD020053); MAM (CA240515) and the NIH through the University of Michigan Rogel Cancer Center Support Grant (P30CA46592). We would like to thank all members of the Zou and Green laboratories for their insightful feedback, as well as L. Fecher. We also acknowledge the support of the University of Michigan Experimental Irradiation Shared Resource, the Immune Monitoring Core, the Vector Core, the University of Michigan Center for Molecular Imaging, the In Vivo Animal Core and the Advanced Genomics Core.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.D.G., J.Y. and W.Z.; Methodology: J.Y. and M.D.G.; Software: S.L., J.E.C., M.C. and C.M.; Formal Analysis: M.C., I.E.N., B.S.R. and Y.S.; Investigation: J.Y., M.D.G., S.L., S.N.J., J.E.C., S.M.R., A.Q., J.F.W., X.L., Z.C., M.S., J.Z., Y.B., L.J., F.S., S.P.N., X.C., S.W., W.S., L.V. and M.C.; Resources: A.Q., C.M., M.M., A.C., C.A.S., K.C., T.S.L., N.R., F.W., M.C., A.A. and W.Z.; Data Curation: A.T., J.S., R.K.A., B.S.R., V.T.M., S.L., M.C., J.J.W. and S.N.J.; Writing — Original Draft: M.D.G., J.Y. and W.Z.; Writing — Review & Editing: M.D.G., J.Y., I.K., N.R., C.D.L. and W.Z.; Supervision: M.D.G., I.K. and W.Z.; Project Administration: J.Y., M.D.G., I.K. and W.Z.; Funding Acquisition: M.D.G., T.S.L., A.M.C., I.E.N., M.A.M, F.W. and W.Z.
Corresponding authors
Ethics declarations
Competing interests
J.Y., M.D.G., S.L., Y.S., S.N.J., J.E.C., S.M.R., J.J.W., X.L., Z.C., J.Z., Y.B., L.J., A.T., J.S., R.K.A., M.S., B.S.R., F.S., S.P.N., X.C., S.W., W.S., L.V., C.M., M.A.M., C.A.S., K.C., I.K., V.T.M., T.S.L., N.R., F.W., A.M.C. and M.C. report no conflicts. A.Q. has research funding from Merck and Clovis. I.E.N. serves as a consultant for Endectra. A.A. serves as a consultant for Merck, AstraZeneca, Bristol-Myers Squibb and Pfizer/EMD Serono. A.A. receives research funding through the University of Michigan from Merck, Genentech, Prometheus Laboratories, Mirati Therapeutics, Roche, Bayer, Progenics, Astellas Pharma, Arcus Biosciences, AstraZeneca, Bristol-Myers Squibb and Clovis Oncology. C.D.L. serves as a consultant for Immunocore. C.D.L receives travel, accommodations and expenses from Bristol-Myers Squibb and Immunocore. C.D.L. receives research funding from Bristol-Myers Squibb, Merck, Novartis and Dynavax. W.Z. has served as a scientific advisor for Cstone, Oncopia and Hengenix.
Additional information
Peer review information Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Liver metastasis correlates with diminished immunotherapy efficacy in cancer patients.
a Best objective response rates in metastatic melanoma patients treated with targeted therapy stratified by baseline disease distribution. Chi-squared for liver metastasis P = 0.63, mean ± SD, liver n = 37, brain n = 23, lung n = 46. b PFS in melanoma patients treated with immunotherapy stratified by liver metastasis. Log-rank test, HR = 2.76, survival±SE, liver n = 64, other n = 118. c OS in melanoma patients treated with targeted therapy stratified by liver metastasis. Log-rank test, HR = 0.557, liver n = 37, other n = 60. d PFS in melanoma patients treated with targeted therapy stratified by liver metastasis; log-rank test, HR = 1.0670, survival±SE, liver n = 37, other n = 60. e Best objective response rates in metastatic NSCLC patients who received chemotherapy stratified by baseline disease distribution. Chi-squared, P = 0.83, mean ± SD, liver n = 43, adrenal n = 39, lung n = 140. f OS in NSCLC patients treated with chemotherapy stratified by liver metastasis; log-rank test, HR = 0.960, survival±SE, liver n = 43, other n = 106. g Forest plot for OS in indicated immunotherapy-treated melanoma patient subset (Cohort 1). Log-rank test, HR, n, and P-value indicated, mean ± SD. h OS in melanoma patients with (n = 55) and without (n = 95) liver metastases treated with immunotherapy in the first-line setting. Log-rank test, HR = 3.564, survival±SE. i OS in melanoma patients with less than the median tumour burden treated with immunotherapy stratified by presence (n = 36) or absence (n = 92) of liver metastasis. Log-rank test, HR = 2.644, survival±SE. j OS in melanoma patients with only liver metastases (n = 15) versus only lung metastases (n = 15). Log-rank test, HR = 3.616; survival±SE. k Forest plot for OS in indicated immunotherapy-treated NSCLC patient subset (Cohort 3). Log-rank test, HR, n, and P-value indicated, mean ± SD. l OS in NSCLC patients treated with immunotherapy in the first line setting stratified by presence (n = 22) or absence (n = 95) of liver metastasis. Log-rank test, HR = 1.577, survival±SE. m OS in NSCLC patients with less than the median tumour burden treated with immunotherapy stratified by presence (n = 25) or absence (n = 115) of liver metastasis. Log-rank test, HR = 2.440; survival±SE. n OS in NSCLC patients wild type EGFR stratified by presence (n = 64) or absence (n = 188) of liver metastasis. Log-rank test, HR = 1.895, survival±SE. o Tumoural PD-L1 staining score in metastatic NSCLC patients (Cohort 3) with (n = 34) and without (n = 114) liver metastases. Unpaired two-tailed Student’s t-test, mean ± SD. p Inverse probability weighted multivariable analysis of PFS in melanoma and NSCLC patients receiving immunotherapy stratified by liver metastasis; Log-rank test, HR = 1.13 and 2.06, respectively, mean ± SD, melanoma liver n = 61, melanoma other n = 102, NSCLC liver n = 63, NSCLC other n = 172. q Variable importance quantification from random forest multivariable modeling of PFS in metastatic melanoma and NSCLC patients receiving immunotherapy (Cohorts 1,3). r Inverse probability weighted multivariable analysis of OS in Cohort 5 stratified by presence of liver metastases. Log-rank test, HR = 2.15, mean ± SD, liver n = 25, other n = 57. s Frequency of relapse in indicated location in melanoma patients with liver metastases receiving immunotherapy. Count displayed. t Frequency of relapse in indicated location in NSCLC patients with liver metastases receiving immunotherapy. Count displayed. u Random effect meta-regression modeling of correlation between anti-PD-1 overall response rates in different histologies versus metastatic tropism to liver or lung. β = 4.6% per 20 liver metastasis increase; β = -0.24% per 20 lung metastasis increase. Mixed effect model, median predicted ORR and 95% confidence intervals, n per Supplementary Table 7.
Extended Data Fig. 2 Liver metastasis diminishes immunotherapy efficacy in mice.
a Schematic for establishing experimental liver metastasis by intrasplenic inoculation. b Schematic for establishing experimental liver tumours by intrahepatic inoculation. c Subcutaneous tumour growth in mice bearing liver tumours established by intrahepatic inoculation, with and without anti-PD-L1 therapy. Two-way ANOVA, mean ± SD, n = 6 per group. d Bioluminescent quantification of secondary tumour burden of a liver tumour from mouse bearing a subcutaneous tumour and liver tumour (as in Fig. 2i) versus the contralateral subcutaneous tumour in mice bearing two subcutaneous tumours (as in Fig. 2h) by bioluminescence. Quantification on day 7 prior to initiation of anti-PD-L1. Unpaired two-tailed Student’s t-test, mean ± SD, n = 10 per group. e, f Subcutaneous MC38 tumour volume (e) and weight (f) in mice with and without liver tumours in which a limited number (1×106) of tumour cells were inoculated subcutaneously. Weight analysed 4 weeks after tumour inoculation. (e) two-way ANOVA; (f) unpaired two-tailed Student’s t-test, mean ± SD, n = 5 per group. Data are representative of at least two independent experiments (c-f).
Extended Data Fig. 3 Liver metastasis induces systemic loss of antigen-specific T cells.
a Subcutaneous tumour CD8+ T cell, IFNγ+CD8+ T cell and Ki67+CD8+ T cell number per gram tumour. Analysed 7 days post anti-PD-L1 treatment initiated. One-way ANOVA with Tukey’s correction, mean ± SD, IgG n = 5, others n = 8. b Representative plot for H-2Kb MuLV p15E tetramer (KSP-tetramer) staining of MC38 tumour-specific CD8+ T cells in S.C. tumour samples. c, d Schematic for unactivated OT-I cell adoptive cell transfer (c) and absolute number of CD45.1+CD8+ OT-I T cells in indicated compartments of mice bearing MC38-Luc or MC38-OVA liver tumour (d). Analysed 14 days after tumour inoculation. Unpaired two-tailed Student’s t-test, mean ± SD, n = 10 per group. e, f Schematic (e) and quantification (f) of activated OT-I-cell distribution 2 days after adoptive cell transfer into mice bearing both subcutaneous and liver MC38-OVA tumour. Displayed as relative cell number to hepatic OT-I cell number. One-way ANOVA, *P = 0.0247, **P = 0.002, ***P = 0.0003, ****P < 0.0001, mean ± SD, n = 6 per group. g, h Representative flow plots (g) and quantification (h) of activated OT-I-cell distribution 4 days after adoptive cell transfer into MC38-OVA tumour bearing mice with MC38-Luc (n = 4) or MC38-OVA (n = 5) liver tumour (as shown in Fig. 3j); unpaired two-tailed Student’s t-test, mean ± SD. i, j Schema (i) and quantification (j) of activated OT-I-cell distribution 4 days after adoptive cell transfer into subcutaneous MC38-OVA tumour bearing mice with MC38-Luc lung tumour or MC38-OVA lung tumour. Displayed as relative cell number (normalized to MC38-Luc group). Unpaired two-tailed Student’s t-test, NS, not significant (P-value: liver, 0.37; lung, 0.42; S.C. tumour, 0.09; spleen, 0.91; tdLN, 0.74; cerLN, 0.26; liverLN, 0.22; lungLN, 0.45; blood, 0.36), mean ± SD, n = 5 per group. k Flow cytometry histogram depicting expression of LFA-1 (left) and CD44 (right) on in vitro activated OT-I cells. l Flow cytometry plots depicting expression of LFA-1 (left) and CD44 (right) expression on in vivo activated OT-I cells isolated from subcutaneous MC38-OVA tumour-draining LNs or non-draining LNs. Analysed 3 days after adoptive transfer. m Flow cytometry histogram depicting expression of LFA-1 (upper) and CD44 (bottom) on tumour specific KSP-tetramer+CD8+ cells (green) in the liver. n CFSE+CD8+OT-I cell number per gram liver tissue. OT-I cells were adoptively transferred 1-2 days after anti-ICAM-1 or HA-se. Analyzed 24 hours after transfer. One-way ANOVA, mean ± SD, control n = 5, HA-se n = 6, anti-ICAM-1 n = 6. o Pre-treatment immune cell subset blood counts of NSCLC patients receiving immunotherapy (Cohort 3) with (n = 62) or without (n = 189) liver metastases. Unpaired two-tailed Student’s t-test, box and whiskers, box represents mean and IQR, whisker represents 10-90%, outliers represent min to max. p Pre-treatment blood immune cell subset counts of NSCLC patients receiving immunotherapy (Cohort 3) with (n = 187) or without (n = 62) lung metastases. Unpaired two-tailed Student’s t-test, box and whiskers, box represents mean and IQR, whisker represents 10-90%, outliers represent min to max. Data are representative of at least two independent experiments (a-n).
Extended Data Fig. 4 T cell phenotype and apoptosis in liver metastasis.
a Flow cytometry histograms showing phenotype of intrahepatic CD45+CD8+KSP-tetramer+ T cell (green) and total CD8 T-cell pool (blue) in mice with subcutaneous MC38 tumours (S.C., bottom) and subcutaneous and liver MC38 tumours (S.C. + liver, top). b Flow cytometry quantification of cleaved caspase-3 of OT-I cells in mice that bearing subcutaneous MC38-OVA tumour and sham (PBS, n = 11), MC38-Luc (n = 10) or MC38-OVA (n = 10) liver tumour. Unactivated CD45.1+CD45.2+OT-I cells were adoptive transferred and analysed 12 days after adoptive transfer. Data from two independent experiments were pooled. One-way ANOVA with Tukey’s correction, mean ± SD. c Frequency of KSP-tetramer+CD8+ cells expressing cleaved caspase-3 in liver of subcutaneous MC38 tumour-bearing mice with (n = 11) and without (n = 6) liver tumours. Unpaired two-tailed Student’s t-test, mean ± SD, data from two independent experiments were pooled. d Cell number of cleaved caspase-3 expressing OT-I cells from indicated location. OT-I cells were activated in vitro and labeled with CFSE, then intravenously transferred. Cells were analysed 4 days after transfer. One-way ANOVA with Dunnett’s multiple comparisons test, P-value: S.C. tumour 0.003, tdLN 0.0045, liverLN 0.0048, cerLN 0.0002, blood 0.0005, mean ± SD, n = 4 per group. e viSNE analysis of indicated marker as detected by CyTOF. Displayed on aggregated samples. Related to Fig. 4c. f Subcutaneous MC38 tumour growth in mice with subcutaneous and liver tumours, treated with anti-PD-L1, anti-CD4, or the combination. Two-way ANOVA with Tukey’s correction, mean ± SD, S.C. +IgG n = 5, S.C. + anti-PD-L1 n = 5, S.C. +liver n = 8, S.C. + anti-PD-L1+ anti-CD4 n = 9. g MC38 subcutaneous tumour growth in mice with subcutaneous and liver tumours, treated with anti-PD-L1, or in combination with hepatic CD4+ adoptive cell transfer (ACT). Two-way ANOVA with Tukey’s correction, mean ± SD, S.C. + IgG n = 9, S.C. + anti-PD-L1 n = 10, S.C. + liver+IgG n = 10, S.C. + liver+anti-PD-L1 n = 8, S.C + liver+CD4 ACT n = 8. Data are representative of at least two independent experiments (a-d).
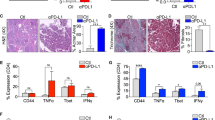
Extended Data Fig. 5 Hepatic myeloid cells induce activated T-cell apoptosis via the Fas/FasL pathway.
a Gating strategy for hepatic CD11b+F4/80+ cells. b Relative cell number of intrahepatic CD11b+F4/80+ following indicated treatment. Samples were analysed after two doses of anti-CSF-1 and clodronate liposome treatment. Data were normalized to control mice receiving PBS liposomes and IgG. One-way ANOVA, mean ± SD, PBS-lipo+IgG n = 9, Clo-lipo+IgG n = 10, PBS-lipo+anti-CSF-1 n = 9, Clo-lipo+anti-CSF-1 n = 11, S.C. n = 8. c Frequency of CD11b+F4/80+ cells (left), absolute number of CD11b+F4/80+ cells (middle) and ratio of CD11b+F4/80+ cells to CD8+ T cells (right) in the liver from mice bearing both MC38 subcutaneous tumour and liver tumour. Samples were collected after two doses of anti-CSF-1 and clodronate liposome treatment. Unpaired two-tailed Student’s t-test, mean ± SD, PBS-lipo+IgG n = 9, Clo-lipo+anti-CSF-1 n = 8. d Absolute number of intrahepatic dendritic cells following two doses of anti-CSF-1 and clodronate liposome treatment. Dendritic cells were gated as CD45+F4/80+CD11c+MHCII+ cells. Unpaired two-tailed Student’s t-test, mean ± SD, PBS-lipo+IgG n = 9, Clo-lipo+anti-CSF-1 n = 8. e Frequency of CD11b+F4/80+ cells (left), absolute number of CD11b+F4/80+ cells (middle) and ratio of CD11b+F4/80+ cells to CD8+ T cells (right) in the subcutaneous tumour from mice bearing both MC38 subcutaneous tumour and liver tumour. Samples were collected after two doses of anti-CSF-1 and clodronate liposome treatment. Unpaired two-tailed Student’s t-test, mean ± SD, n = 7 per group. f MC38 subcutaneous tumour growth in mice with only S.C. tumours treated with anti-PD-L1, clodronate liposome and anti-CSF-1, or the combination. Two-way ANOVA with Tukey’s correction, mean ± SD, n = 8 per group. g Schematic for clodronate liposome, anti-CSF-1, and OT-I adoptive transfer. h MC38 subcutaneous tumour growth in mice with S.C. and liver tumours treated with anti-PD-L1, clodronate liposome, anti-CSF-1, anti-CD8, or the combination. Two-way ANOVA with Tukey’s correction, mean ± SD, n = 6 per group. i Frequency of annexin V+7-AAD+ OT-I cells co-cultured in the presence of OVA peptide with hepatic F4/80+ cells isolated from liver tumour bearing mice at indicated ratios for 48 hours; Activated OT-I cells were labeled with CFSE before co-culture. One-way ANOVA with Dunnett’s multiple comparisons test, mean ± SD, n = 7 biologically independent samples. j Frequency of annexin V+7-AAD+ OT-I cells (CFSE labeled) after co-cultured in the presence of OVA peptide with hepatic F4/80+ cells in indicated conditions for 48 hours. One-way ANOVA with Tukey’s correction, mean ± SD, n = 3 biologically independent samples. k Flow cytometry histogram of Fas expression on hepatic OT-I (left) and KSP-tetramer+CD8+ T cells (right). Unactivated OT-I cells were transferred into mice bearing MC38-OVA subcutaneous tumour and liver tumour. Phenotype of transferred OT-I cells and endogenous KSP-tetramer+CD8+ T cells were analysed 12 days after adoptive transfer. l Frequency of annexin V+7-AAD+ OT-I cells co-cultured with MC38-OVA tumour cells and hepatic F4/80+ cells isolated from liver tumour bearing mice with and without TNFα blockade. Activated OT-I cells were labeled with CFSE before co-culture. Unpaired two-tailed Student’s t-test, mean ± SD, n = 4 biologically independent samples. m Quantification of H-2Kb-OVA mean fluorescent intensity (MFI) on hepatic CD11b+F4/80+ cells recovered from mice bearing subcutaneous MC38-OVA tumour with or without liver MC38-OVA tumour. Unpaired two-tailed Student’s t-test, mean ± SD, n = 5 per group. n, o Quantification of FasL (n) and H-2Kb (o) MFI on lung CD11b+F4/80+ cells recovered from mice bearing subcutaneous MC38 tumour with (n = 9) or without (n = 6) lung MC38 tumour, in comparison with hepatic CD11b+F4/80+ cells recovered from mice bearing subcutaneous MC38 tumour with (n = 9) or without (n = 6) liver MC38 tumour. Tissues were collected 10 days after tumour inoculation. One-way ANOVA, mean ± SD. Data are representative of at least two independent experiments (b-o).
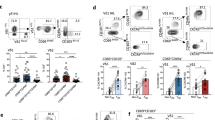
Extended Data Fig. 6 Liver metastasis alters the liver immune microenvironment.
a UMAP plot of all hepatic immune cell clusters on all samples merged. b Table identifying immune cell clusters and listing key genes. c Frequency of all immune cell clusters in mice with a subcutaneous tumour (S.C.) and mice with subcutaneous and liver tumours (S.C. + liver). d Apoptosis gene set enrichment analysis of in activated T-cell clusters in mice with a subcutaneous tumour and mice with subcutaneous and liver tumours. Activated T cells were identified by expression of Cd44. Unpaired two-tailed Student’s t-test, min to max, S.C. n = 157 cells, S.C. + liver n = 38 cells. e UMAP plot of Lgal3 on all samples merged. f UMAP plot of residential and migratory macrophages on all samples merged. g Violin plot of residential and migratory macrophage gene signatures in mice with a subcutaneous tumour and mice with subcutaneous and liver tumours. Unpaired two-tailed Student’s t-test, min to max. h Violin plot of residential macrophage M2-like and M1-like gene set enrichment in mice with a subcutaneous tumour and mice with subcutaneous and liver tumours. Unpaired two-tailed Student’s t-test, NS, not significant, min to max. S.C. n = 543 cells, S.C. + liver n = 936 cells. i Violin plot of cross-presentation gene set enrichment in resident (n = 1479 cells) and mono-derived (n = 6698 cells) macrophages. Unpaired two-tailed Student’s t-test, min to max. j Violin plot of M2-like and M1-like signatures within monocyte-derived macrophage cell subsets in mice with a subcutaneous tumour and mice with subcutaneous and liver tumours. Unpaired two-tailed Student’s t-test, min to max. k Pseudotime analysis overlying time with monocyte-derived macrophage cell states. l Pseudotime analysis overlaying monocyte-derived macrophage clusters and states.
Extended Data Fig. 7 Extended CyTOF data analysis of liver immune cells after radiotherapy and immunotherapy.
a Schematic describing combination treatment with liver directed radiotherapy followed by anti-PD-L1. b-d Immune clusters identified by mass cytometry in Fig. 6a. b, Heatmap showing frequency of antibody labeling (rows) in the 18 immune populations (columns) derived from a combined analysis of all samples. c, Immune cell subset identified by mass cytometry. d, viSNE representation of key marker expression across subpopulations, displayed on aggregated samples.
Extended Data Fig. 8 Radiotherapy reshapes the liver immune microenvironment.
a, b Flow cytometry quantification of CD8+ T-cell number (a), Ki67+, IFNγ+ and granzyme B+ CD8+ T cells (b) in the livers from mice bearing both subcutaneous tumour and liver tumour with indicated treatments. Analysed 5 days after RT. One-way ANOVA, mean ± SD, n = 6 per group. c Hepatic tissue chemokine levels in subcutaneous and liver tumour bearing mice treated in indicated fashion. Analysed 5 days after RT by Luminex; One-way ANOVA, mean ± SD, n = 5 per group. d-f ELISA analysis of culture supernatant (d, n = 3 biologically independent samples) and phenotypic analysis (e, f, n = 4 biologically independent samples) of MC38 cells 48 hours after irradiation. One-way ANOVA, **P = 0.006, ***P = 0.0003, ****P < 0.0001, mean ± SD. g Flow cytometry plot and quantification of cleaved caspase-3 in total hepatic CD8+ T cells from mice bearing both subcutaneous tumour and liver tumour, with (n = 8) or without (n = 7) liver-directed radiotherapy (RT). Unpaired two-tailed Student’s t-test, mean ± SD. h Schematic describing liver-directed radiotherapy and adoptive cell transfer. Data are representative of at least two independent experiments (a-g).
Extended Data Fig. 9 Radiotherapy abolishes immunotherapy resistance induced by liver metastasis.
a, b Flow cytometry quantification of subcutaneous tumour dLN Ki67+ (a) and IFNγ+ (b) in CD8+ and CD4+ T cells in mice with subcutaneous and liver metastasis treated as indicated; analysed 7 days after radiotherapy. One-way ANOVA, mean ± SD, IgG n = 4, others n = 5. c Representative bioluminescent imaging of subcutaneous and liver tumour bearing mice following treatment with anti-PD-L1, radiotherapy, and anti-CD8. d Subcutaneous tumour volume of mice bearing only subcutaneous MC38 tumours treated with radiation to the liver, anti-PD-L1, or the combination. Two-way ANOVA with Tukey’s correction, mean ± SD, n = 7 per group. e Subcutaneous tumour volume of mice bearing subcutaneous KPC2 tumours treated with anti-PD-L1. Two-way ANOVA, mean ± SD, n = 10 per group. f KPC2 subcutaneous tumour growth in mice with (S.C. +liver) or without (S.C.) liver tumours treated with anti-PD-L1, or in combination with liver directed radiotherapy. f, Two-way ANOVA, mean ± SD with Tukey’s correction, n = 10 per group. g KPC2 liver tumour growth in mice with subcutaneous and liver tumours treated as in (f). One-way ANOVA, mean ± SD, n = 5 for IgG and anti-PD-L1 group, n = 7 for anti-PD-L1+RT group. Data are representative of at least two independent experiments (a-g).
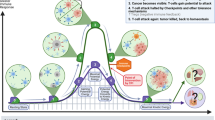
Extended Data Fig. 10 Impact of liver metastasis on cancer-immunity cycle.
The graphical abstract describes how liver metastases alter the normal cancer immunity cycle by inducing hepatic siphoning of T cells, and further how liver-directed radiotherapy can disrupt hepatic siphoning to promote effective anti-tumoral immunity.
Supplementary information
Supplementary Information
Supplementary Tables 1–8 and Supplementary Figs. 1–8.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Yu, J., Green, M.D., Li, S. et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med 27, 152–164 (2021). https://doi.org/10.1038/s41591-020-1131-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-1131-x
This article is cited by
-
Potential crosstalk between SPP1 + TAMs and CD8 + exhausted T cells promotes an immunosuppressive environment in gastric metastatic cancer
Journal of Translational Medicine (2024)
-
Progress of immune checkpoint inhibitors therapy for non-small cell lung cancer with liver metastases
British Journal of Cancer (2024)
-
Class II HLA-DRB4 is a predictive biomarker for survival following immunotherapy in metastatic non-small cell lung cancer
Scientific Reports (2024)
-
Pirfenidone alleviates fibrosis by acting on tumour–stroma interplay in pancreatic cancer
British Journal of Cancer (2024)
-
Serum amyloid A promotes glycolysis of neutrophils during PD-1 blockade resistance in hepatocellular carcinoma
Nature Communications (2024)