Abstract
We present cleared-tissue axially swept light-sheet microscopy (ctASLM), which enables isotropic, subcellular resolution imaging with high optical sectioning capability and a large field of view over a broad range of immersion media. ctASLM can image live, expanded, and both aqueous and non-aqueous chemically cleared tissue preparations. Depending on the optical configuration, ctASLM provides up to 260 nm of axial resolution, a three to tenfold improvement over confocal and other reported cleared-tissue light-sheet microscopes. We imaged millimeter-scale cleared tissues with subcellular three-dimensional resolution, which enabled automated detection of multicellular tissue architectures, individual cells, synaptic spines and rare cell–cell interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The datasets acquired for this study are available from the corresponding authors upon reasonable request.
Code availability
The instrument control software can be requested for academic use from the corresponding authors and will be delivered under material transfer agreements with Howard Hughes Medical Institute and UT Southwestern Medical Center.
References
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes and progenitors. Science 356, eaah4573 (2017).
Wang, K. et al. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat. Methods 11, 625–628 (2014).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Welf, ErikS. et al. Quantitative multiscale cell imaging in controlled 3D microenvironments. Dev. Cell 36, 462–475 (2016).
Huisken, J. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Dodt, H.-U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Stefaniuk, M. et al. Light-sheet microscopy imaging of a whole cleared rat brain with Thy1–GFP transgene. Sci. Rep. 6, 28209 (2016).
Pende, M. et al. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 9, 4731 (2018).
Tomer, R. et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163, 1796–1806 (2015).
Buytaert, J. A. & Dirckx, J. J. Tomographic imaging of macroscopic biomedical objects in high resolution and three dimensions using orthogonal-plane fluorescence optical sectioning. Appl. Opt. 48, 941–948 (2009).
Santi, P. A. et al. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. Biotechniques 46, 287–294 (2009).
Dean, KevinM. et al. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophysical J. 108, 2807–2815 (2015).
Botcherby, E. J. et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl Acad. Sci. USA 109, 2919–2924 (2012).
Gao, L. Extend the field of view of selective plan illumination microscopy by tiling the excitation light sheet. Opt. Express. 23, 6102–6111 (2015).
Voigt, F. F. et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat. Methods https://doi.org/10.1038/s41592-019-0554-0 (2019).
Hörl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019).
Gao, R. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019).
Fu, Q., Martin, B. L., Matus, D. Q. & Gao, L. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat. Commun. 7, 11088 (2016).
Crane, G. M., Jeffery, E. & Morrison, S. J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590 (2017).
Driscoll, M. K. et al. Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat. Methods 16, 1037–1044 (2019).
Acar, M. et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015).
Comazzetto, S. et al. Restricted hematopoietic progenitors and erythropoiesis require scf from leptin receptor + niche cells in the bone marrow. Cell Stem Cell 24, 477–486 (2018).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Migliori, B. et al. Light-sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 16, 57 (2018).
Guenthner, CaseyJ. et al. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784 (2013).
Jing, D. et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 28, 803–818 (2018).
Sorensen, I., Adams, R. H. & Gossler, A. DLL1-mediated notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680–5688 (2009).
Xu, W. & Sudhof, T. C. A neural circuit for memory specificity and generalization. Science 339, 1290–1295 (2013).
Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Schindelin, J. et al. Fiji: an open source platform for biological image analysis. Nat. Methods 9, 676–682 (2012).
Applegate, K. T. et al. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176, 168–184 (2011).
Cignoni, P. et al. MeshLab: an open source mesh processing tool. In Proc. Eurographics Italian Chapter Conference (eds Scarano, V. et al) 129–136 (Eurographics Association, 2008).
Acknowledgements
We thank the Cancer Prevention Research Institute of Texas (grant nos. RR160057 to R.F. and R1225 to G.D.) for their generous funding, as well as the National Institutes of Health (grant nos. F32GM116370 and K99GM123221 to M.K.D., R01GM067230 to G.D., R01AG055577 and R01NS056224 to I.B., R01DK118032 and R01DK099478 to D.M., R01DC015784 and R21NS104826 to J.M., R33CA235254 and R35GM133522 to R.F., and DP2MH119423 to R.T.). R.H. acknowledges support by the Collaborative Research Center SFB 1278 (PolyTarget, project C04) funded by the Deutsche Forschungsgemeinschaft. We are grateful to the Live cell imaging facility at UT Southwestern for access to the Zeiss Airyscan confocal microscope (supported by grant no. 1S10OD021684-01). We thank D. Saucier and J. Amatruda for providing the zebrafish specimens and E. Sapoznik for his assistance with computer-aided design.
Author information
Authors and Affiliations
Contributions
T.C., K.M.D. and R.F. designed the research. T.C. and R.F designed and built the microscopes. T.C. and S.V. prepared the CAD files and operated the microscope. T.C., M.K.D., K.M.D., B-J.C, P.R. and R.F. performed image analysis, partially under guidance by G.D. R.H. simulated PSFs. W.M.W., C.N., H.Z., V.Z., M.M.M., E.J., C.H., D.M., I.B., H.Z., R.T., J.M. and S.M. provided specimens and guided imaging. T.C., K.M.D. and R.F. wrote the manuscript. All authors read and provided feedback on the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Simulations for the effect of axially scanning the light-sheet only with defocus, as it would occur with electro-tunable lenses.
(a) Cross-section of a light-sheet with NA 0.4 at nominal focal plane of the excitation objective. (b) Cross-section of a light-sheet with NA 0.4 defocused by 500 um. (c) Cross-section of a light-sheet with an NA of 0.7 at nominal focus. (d) Cross-section of a light-sheet with NA 0.7 defocused by 150 um. Scale bar: 5 um.
Supplementary Figure 2 PSF of ctASLM over a refractive index range of 1.33-1.54.
XY and XZ views of the PSFs as shown in water (RI~1.33), 54% glycerol (RI~1.42) and PEGASOS (RI~1.543). PSFs were generated by using 200 nm green fluorescent beads. Scale bar: 2 μm (ctASLM1, NA 0.4) and 1 μm (ctASLM2, NA 0.7).
Supplementary Figure 3 ctASLM allows isotropic, sub-micron imaging over a wide refractive index range.
(a) Vasculature of a live 72-hour old zZebrafish imaged in E3 medium. Vasculature endothelial cells labeled with mCherry. The inset shows a zoomed in view of the heart. Scale bar: 500 µm. (b) Thy1-eYFP neurons in a mouse brain cleared with CLARITY. Scale bar: 500 µm. (c) Segmentation of two neurons (magenta and green) and a dendrite from a neighboring neuron (yellow) from the dataset shown in (a), Scale bar: 50 µm. Insets shows magnified views of dendrites and their spines. (d) Volume rendering of a mouse kidney labeled with Flk1-GFP. Scale bar: 500 µm. (e) one cross-section through the kidney shown in (d), Scale bar: 500 µm. Inset shows a 3D rendering of endothelial cells in one selected Glomeruli.
Supplementary Figure 4 XZ view over 870×870 µm2 lateral field of view depicting PSFs with sub-micron isotropic resolution.
PSFs were generated using 200 nm green fluorescent beads embedded in a 2% agarose cube, no deconvolution was applied to the shown data. The resolutions (n= 5, mean ± standard deviation) are 0.9 ± 0.03 µm laterally and 0.85 ± 0.03 µm axially, respectively. Scale bar: 100 µm and 5 µm, respectively.
Supplementary Figure 5 XZ view over 320×320 µm2 field of view depicting PSFs with sub-micron isotropic resolution.
PSFs were generated using 200 nm green fluorescent beads embedded in a 2% agarose cube, no deconvolution was applied to the shown data. The resolutions (n= 5, mean ± standard deviation) are 480± 12 nm laterally and 480±29 nm axially, respectively. Scale bar: 50 µm and 2 µm, respectively.
Supplementary Figure 6 Blind deconvolution can improve spatial resolution by ~30%.
Beads are embedded in 2% agarose and submersed in water. The images were acquired with ctASLM1 microscope with NA 0.4 objectives in both illumination and detection. (a,c) X-Y maximum intensity projection of 200 nm fluorescent beads before and after 5 rounds of blind deconvolution, respectively. (b,d) X-Z maximum intensity projection before and after blind deconvolution, respectively. We measured 10 isolated beads and the average resolution (n=10, mean ± standard deviation) is 950 ± 30 nm (xy) and 950 ± 115 nm (z) before deconvolution, and 770 ± 33 nm (xy) and 750 ± 87 nm (z) after deconvolution. The data shown was resampled by 2x.
Supplementary Figure 7 Images of beads before and after blind deconvolution.
Deconvolution can improve spatial resolution by ~30% both laterally and axially. The sample consists of 200 nm beads embedded in 2% agarose and immersed in water. The images were acquired with ctASLM2 microscope with NA 0.7 objectives in both illumination and detection. With such high NA objectives, we measured 10 isolated beads and the average resolution (n=10, mean ± standard deviation) is 479 ± 11.7 nm (xy) and 483 ± 29.1 nm (z) before deconvolution, and 358 ± 12.3 nm (xy) and 366 ± 17.8 nm (z) after deconvolution. The improvement is 1.32x laterally and 1.34x axially, respectively. The data shown here was re-sampled by 2x.
Supplementary Figure 8 Quantifying the resolution in a cleared tissue.
Bone marrow cleared with BABB, nerve fibers labeled with Alexa 488. Images have been deconvolved with blind deconvolution with 10 iterations. Small clumps of antibodies in the specimen are used to quantify the resolution. (a) Maximum intensity projection (MIP) of the whole stack in xy. Three arrows in (a) show examples of small antibody clumps. (b) Ten clumps were selected for quantifying the resolution. Images show the MIPs in xy, zy, and xz views. The scale bar is 1 µm. The resolution averaged from these clumps of antibodies is (n= 10, mean ± standard deviation) 311 ± 22.8 nm laterally and 287 ± 28.0 nm axially, respectively.
Supplementary Figure 9 Comparison of the images before (left column) and after Blind deconvolution (right column).
Thy1 GFP Neurons cleared with PEGASOS imaged by ctASLM. (a,c) X-Y maximum intensity projection before and after 10 rounds of deconvolution, respectively. (b,d) X-Z maximum intensity projection before and after deconvolution, respectively. The data was resampled by 2x bicubic interpolation in ImageJ prior to the deconvolution. Gamma correction with 0.3 and 0.5 was applied on non-deconvolved and deconvolved images, respectively to balance the brightness between the spines and the main cell body of neurons.
Supplementary Figure 10 Isotropic, sub-micron imaging of Thy1-eYFP neurons in a mouse brain cleared with PEGASOS.
(a) Maximum intensity projections (MIP) in xy of the whole mouse brain stitched from stacks at one z position (same region as in Fig. 2b, layer is located at 2.5 mm deep from the surface). The full size of the MIP is about 10.7 mm × 8.8 mm. (b1)-(b6) MIPs in xy, zy, and xz of the selected regions 1-6 shown in (a). The size of individual volume is 737 µm × 737 µm × 450 µm. The images were corrected with gamma at a value of 0.5 to balance the brightness.
Supplementary Figure 11 Zoom in of the image shown in Supplementary Fig. 10.
(a) The same image as shown in Supplementary Fig. 10 (b1). (b1-b8) Magnified views of the selected regions 1-8 shown in (a). Note that (b1)-(b8) are from different regions in either the XY, YZ, or XZ maximum intensity projections. Arrows in the insets (b2), (b4), and (b8), point the spines.
Supplementary Figure 12 Analysis of detected spines.
(Top) Variation from a sphere. (Bottom) Ratio of spine neck surface area to total spine surface area. Colors correspond to clusters identified by hierarchical clustering, and both x-axes are unitless. This experiment was performed once (n=1).
Supplementary Figure 13 Detected endothelial cells in a glomeruli.
Individual glomeruli were identified and masked using a watershed filter on data that was filtered with a 20-pixel 3D Gaussian blur. Cells within a glomerulus were then detected within the masked area using a second watershed algorithm. Scale bar: 50 µm.
Supplementary Figure 14 High resolution image of nerve (green) and cKit labeled cells (magenta) in a Pegasos cleared bone marrow.
(a) Maximum intensity projections (MIPs) of the whole stack in XY, YZ, and XZ. The full size of the stack is 267.2 µm × 267.2 µm × 165.2 µm. Zoom in of boxes 1, 2, 3, and 4 are shown in b1, b2, b3, and b4, respectively. (b1)-(b4) MIPs in xy, zy, and xz of selected regions shown in (a). The size of the selected volumes is 40 µm × 40 µm × 45 µm. Note that MIPs are taken through a shorter depth in z, allowing us to see more clearly some of the details. (c) Zoom in of the box 4’ shown in (b4), white arrows point at twisted nerve fibers. Image data was acquired with ctASLM2 (NA 0.7) and has been deconvolved with blind deconvolution with 10 iterations.
Supplementary Figure 15 Imaging of cKit+ cells in a PEGASOS cleared bone marrow.
(a) Cross-sectional view acquired with ctASLM2 (NA 0.7). (b) Cross-sectional view acquired with a confocal microscope with a Leica BABB objective with an NA of 0.9. (c) Cross-sectional view acquired with a Zeiss Airy scan confocal with an NA of 0.8. (d-i) cross-sectional views acquired with ctASLM2, showing hot spots of cKIT labeling and small membrane protrusions. Scale bar: 5 µm.
Supplementary Figure 16 Maximum intensity projection of a chemically expanded hippocampal slice.
Expanded hippocampal slice from a Thy1-GFP mice as imaged with ctASLM1 equipped with NA 0.4 objectives. Insets show magnified views of the boxed regions. Scale bar 100 microns (includes magnification by expansion and by the microscope).
Supplementary Figure 17 Maximum intensity projection of expanded neurons.
Maximum intensity projection of mCherry-expressing AAV-Synaptotag transduced neurons from a chemically expanded hippocampal slice as imaged with ctASLM equipped with NA 0.7 objectives. Scale bar 5 microns (includes magnification by expansion and by the microscope).
Supplementary Figure 18 Maximum intensity projection of a chemically expanded hippocampal slice.
Maximum intensity projections from a Thy1-GFP mice (green for GFP, presynaptic terminal) and post synaptic density labeled with anti-Homer1 antibodies (magenta) imaged with ctASLM using NA 0.7 objectives. XY and XZ image projections are shown for two different synapses from the same hippocampal sample. Scale bar: 1 µm (accounting for microscope magnification and expansion).
Supplementary Figure 19 Depth performance of ctASLM within tissues.
(a) mouse kidney shown in Fig. 1g. White arrows point to the location where the kidney was glued to the sample holder. Scale bar 500 µm. (b-c) Magnified views of the green boxed area in (a). Scale bar: 20 µm. (d-e) Magnified view of the blue boxed area in (a). (f) Schematic illustration of the illumination geometry. (g) Slice of a data volume acquired from a Clarity cleared mouse brain. (h-i) Magnified views of the boxed yellow area in (g). (j-k) Magnified view of the red boxed area in (g). (l) Location of the slice relative to the illumination and detection objective. Scale bar: 20 µm.
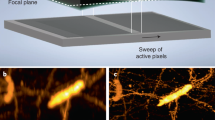
Supplementary Figure 20 Schematic drawing (top) and CAD rendering (bottom) of the ctASLM system using NA 0.4 objectives.
A part list is provided in Supplementary Table 4 for the ctASLM version using NA 0.4 objectives.
Supplementary Figure 21 Compensation of spherical aberrations in light-sheets.
The remote focusing arm of ctASLM 1 (NA 0.4) uses an Olympus objective that is designed to image with a working distance that consists of 24 mm of air and 5 mm of water. Here, the objective was used with only an air interface, simplifying the operation of the remote focusing system. By introducing a 3 mm of glass window in the image space, spherical aberrations were eliminated. (a) and (b) show the light sheets with and without the spherical aberration by adding and removing the glass coverslip, respectively. Scale bar: 50 µm.
Supplementary Figure 22 Schematic drawing of the ctASLM system using NA 0.7 objectives.
A part list for ctASLM using the NA 0.7 objectives is provided in Supplementary Table 5.
Supplementary Figure 23 Mounting of the sample and custom sample holder assembly.
Main figure: CAD rendering of the sample mount and the adaptor to the sample scanning stage. (a) A neonatal mouse kidney is glued to the glass slide using superglue. (b) Bone marrow embedded in cleared agarose. The agarose-cylinder is then held to the glass slide using silicone form the two ends.
Supplementary Figure 24 Comparison of imaging performance between ctASLM and a confocal microscope.
Cortex of a PEGASOS cleared mouse brain expressing THy1-GFP (a) XY and (b) YZ were acquired using ctASLM using NA 0.4 objectives. (c) and (d) are a representative area imaged using a confocal microscope. Scale bar: 25 µm.
Supplementary Figure 25 Calibration of the scan range using a 2D laser focus in fluorescein.
XY image in light-sheet mode while a 2D laser focus is scanned across the image via remote focusing and synchronizing the scan motion to the light-sheet mode readout of the camera. The scanned laser focus shows up as a white line in the center of the camera frame. If the scan range and relative phase between scan motion and camera readout matches, a think line appears. If the scan range does not match, the line becomes less uniform and less sharp.
Supplementary information
Supplementary Information
Supplementary Figs. 1–25, Notes 1–4 and Tables 1–7.
Supplementary Video 1
Neuron segmentation: segmented Thy1-GFP cortical neurons from a CLARITY-cleared mouse brain in a subregion spanning 170 × 170 × 60 μm3.
Supplementary Video 2
Large-scale brain imaging with ctASLM: volume rendering of PEGASOS-cleared mouse brain labeled with Thy1-GFP.
Supplementary Video 3
Close-up of brain data: close-up of a maximum-intensity projection of a section of PEGASOS-cleared mouse brain.
Supplementary Video 4
Rendering of a segmented neuron: segmentation of a neuron in a PEGASOS-cleared mouse brain. Coordinate axes measure 50 μm.
Supplementary Video 5
Kidney imaging with ctASLM: BABB-cleared mouse kidney labeled with Flk1-GFP in an imaging volume of 3.4 × 2.6 × 2.5 mm3.
Supplementary Video 6
Stem cell imaging with ctASLM and Airyscan imaging of HSCs (gray) and niche cells (red) using ctASLM (left) and Airyscan (right). Labeling: lepR+ niche cells (tdTomato) and c-Kit+ cells (HSCs; Alexa Fluor 647). Each imaging volume measures 40 × 40 × 20 μm3.
Supplementary Video 7
Olfactory bulb imaging with ctASLM: olfactory bulb neurons are labeled with a stimulus-inducible tdTomato reporter and cleared with PEGASOS. Imaging region spans 2.8 × 2 × 0.66 mm3.
Supplementary Video 8
Curvature of spines: detection of spines on Thy1-eYFP neurons on the basis of curvature over a 176 × 190 × 60 μm3 volume.
Supplementary Video 9
Clustering of spines: clustering of spines following principal-component analysis of their morphological parameters. Volume spans 176 × 190 × 60 μm3.
Supplementary Video 10
Glomeruli detection: automatically detected glomeruli from a mouse kidney labeled with Flk1-GFP. Scale bar, 25 microns.
Supplementary Video 11
Bone marrow imaging: volume rendering of a subvolume of a cleared bone marrow sample. Sinusoids are rendered white, arterioles are orange, nerve fibers are green and progenitor cells are magenta. Blood vessels are labeled with tdTomato, Nerve fibers are labeled with Alexa 488, progenitor cells are labeled with Alexa 647. Coordinate axes measure 10 μm each.
Supplementary Video 12
Expansion microscopy: Thy1-GFP neuron labeled with mCherry from a chemically expanded hippocampal slice imaged with ctASLM equipped with NA 0.7 objectives. Imaging volume is 300 × 155 × 126 μm3.
Supplementary Video 13
Expansion microscopy: z-stack depicting mCherry-labeled Thy1-GFP neurons from a chemically expanded hippocampal slice imaged with ctASLM equipped with NA 0.7 objectives.
Rights and permissions
About this article
Cite this article
Chakraborty, T., Driscoll, M.K., Jeffery, E. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat Methods 16, 1109–1113 (2019). https://doi.org/10.1038/s41592-019-0615-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0615-4
This article is cited by
-
Signal improved ultra-fast light-sheet microscope for large tissue imaging
Communications Engineering (2024)
-
Current and future applications of light-sheet imaging for identifying molecular and developmental processes in autism spectrum disorders
Molecular Psychiatry (2024)
-
Benchtop mesoSPIM: a next-generation open-source light-sheet microscope for cleared samples
Nature Communications (2024)
-
Efficient 3D light-sheet imaging of very large-scale optically cleared human brain and prostate tissue samples
Communications Biology (2023)
-
Intestinal epithelial barrier integrity investigated by label-free techniques in ulcerative colitis patients
Scientific Reports (2023)