Abstract
The ability to directly measure acetylcholine (ACh) release is an essential step toward understanding its physiological function. Here we optimized the GRABACh (GPCR-activation-based ACh) sensor to achieve substantially improved sensitivity in ACh detection, as well as reduced downstream coupling to intracellular pathways. The improved version of the ACh sensor retains the subsecond response kinetics, physiologically relevant affinity and precise molecular specificity for ACh of its predecessor. Using this sensor, we revealed compartmental ACh signals in the olfactory center of transgenic flies in response to external stimuli including odor and body shock. Using fiber photometry recording and two-photon imaging, our ACh sensor also enabled sensitive detection of single-trial ACh dynamics in multiple brain regions in mice performing a variety of behaviors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The plasmids pAAV-hsyn-ACh3.0 and pAAV-hsyn-DIO-ACh3.0 have been deposited to Addgene (121922 and 121933) and are available. The transgenic Drosophila and related materials have been deposited to the Bloomington Drosophila Stock Center (flies: FBti0210605, FBti0210606, FBti0210607 and FBti0210608; plasmids: FBtp0140745, FBtp0140746, FBtp0140747 and FBtp0140748) and are available. Source data are provided with this paper.
Code availability
The custom-written MATLAB, Arduino and TDT programs will be provided upon request.
References
Dale, H. H. The action of certain esters and ethers of choline, and their relation to muscarine. J. Pharmacol. Exp. Ther. 6, 147–190 (1914).
Hasselmo, M. E. The role of acetylcholine in learning and memory. Curr. Opin. Neurobiol. 16, 710–715 (2006).
Winkler, J., Suhr, S., Gage, F., Thal, L. & Fisher, L. Essential role of neocortical acetylcholine in spatial memory. Nature 375, 484–487 (1995).
Krnjević, K. & Miledi, R. Acetylcholine in mammalian neuromuscular transmission. Nature 182, 805–806 (1958).
Magleby, K. L. & Stevens, C. F. A quantitative description of end-plate currents. J. Physiol. 223, 173–197 (1972).
Marrosu, F. et al. Microdialysis measurement of cortical and hippocampal acetylcholine release during sleep–wake cycle in freely moving cats. Brain Res. 671, 329–332 (1995).
Nguyen, Q.-T. et al. An in vivo biosensor for neurotransmitter release and in situ receptor activity. Nat. Neurosci. 13, 127–132 (2010).
Jing, M. et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol. 36, 726–737 (2018).
Zhang, X., Noyes, N. C., Zeng, J., Li, Y. & Davis, R. L. Aversive training induces both presynaptic and postsynaptic suppression in Drosophila. J. Neurosci. 39, 9164–9172 (2019).
Zhu, P. K. et al. Nanoscopic visualization of restricted nonvolume cholinergic and monoaminergic transmission with genetically encoded sensors. Nano Lett. 20, 4073–4083 (2020).
Tautermann, C. S. et al. Molecular basis for the long duration of action and kinetic selectivity of tiotropium for the muscarinic M3 receptor. J. Med. Chem. 56, 8746–8756 (2013).
Disse, B. et al. Ba 679 BR, a novel long-acting anticholinergic bronchodilator. Life Sci. 52, 537–544 (1993).
Falkenburger, B. H., Jensen, J. B. & Hille, B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J. Gen. Physiol. 135, 81–97 (2010).
Ziegler, N., Bätz, J., Zabel, U., Lohse, M. J. & Hoffmann, C. FRET-based sensors for the human M1-, M3-, and M5-acetylcholine receptors. Bioorg. Med. Chem. 19, 1048–1054 (2011).
Wan, Q. et al. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J. Biol. Chem. 293, 7466–7473 (2018).
Barnea, G. et al. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69 (2008).
Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5, e12727 (2016).
Zhang, J. et al. Presynaptic excitation via GABAB receptors in habenula cholinergic neurons regulates fear memory expression. Cell 166, 716–728 (2016).
Yao Yang, M., Armstrong, J. D., Vilinsky, I., Strausfeld, N. J. & Kaiser, K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron 15, 45–54 (1995).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Vazquez, J. & Baghdoyan, H. A. Basal forebrain acetylcholine release during REM sleep is significantly greater than during waking. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R598–R601 (2001).
Picciotto, M. R., Higley, M. J. & Mineur, Y. S. Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron 76, 116–129 (2012).
Jiang, L. et al. Cholinergic signaling controls conditioned fear behaviors and enhances plasticity of cortical–amygdala circuits. Neuron 90, 1057–1070 (2016).
Li, X. et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl Acad. Sci. USA 115, 415–420 (2018).
Seltzer, B. Donepezil: a review. Expert Opin. Drug Metab. Toxicol. 1, 527–536 (2005).
Martins-Silva, C. et al. Novel strains of mice deficient for the vesicular acetylcholine transporter: insights on transcriptional regulation and control of locomotor behavior. PLoS ONE 6, e17611 (2011).
Xu, M. et al. Basal forebrain circuit for sleep–wake control. Nat. Neurosci. 18, 1641–1647 (2015).
Cheung, J., Maire, P., Kim, J., Sy, J. & Hires, S. A. The sensorimotor basis of whisker-guided anteroposterior object localization in head-fixed mice. Curr. Biol. 29, 3029–3040 (2019).
Zong, W. et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017).
Patriarchi, T. et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018).
Feng, J. et al. A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 102, 745–761 (2019).
Liang, R., Broussard, G. J. & Tian, L. Imaging chemical neurotransmission with genetically encoded fluorescent sensors. ACS Chem. Neurosci. 6, 84–93 (2015).
Jing, M., Zhang, Y., Wang, H. & Li, Y. G-protein-coupled receptor-based sensors for imaging neurochemicals with high sensitivity and specificity. J. Neurochem. 151, 279–288 (2019).
Sun, F. et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481–496 (2018).
Al-Onaizi, M. A. et al. Regulation of cognitive processing by hippocampal cholinergic tone. Cereb. Cortex 27, 1615–1628 (2017).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Janickova, H. et al. Selective decrease of cholinergic signaling from pedunculopontine and laterodorsal tegmental nuclei has little impact on cognition but markedly increases susceptibility to stress. FASEB J. 33, 7018–7036 (2019).
Acknowledgements
We thank Y. Rao (Peking University) for generously sharing a two-photon microscope and X. Lei (Peking University) for providing support of the Opera Phenix High-Content Screening System at PKU–CLS. We thank W. Inoue (University of Western Ontario) for kindly sharing the Multi Conditioning System foot-shocker. Work at the University of Western Ontario was supported by the Canada First Research Excellence Fund (CFREF; to L.M.S., V.F.P., T.J.B. and M.A.M.P.) and CIHR (PJT 159781 to V.F.P.). O.K. was supported by an OGS PhD Fellowship and the Jonathan and Joshua Graduate Scholarship. M.S. was supported by CFREF. This work was supported by the General Research Program of the National Natural Science Foundation of China (projects 31671118 to Yulong Li and 31327901 to H.C.), the NIH BRAIN Initiative (grant U01NS103558 to Yulong Li), the Beijing Brain Initiative of the Beijing Municipal Science & Technology Commission (Z181100001518004 to Yulong Li), the Junior Thousand Talent Program of China and grants from the Peking–Tsinghua Center for Life Sciences and the State Key Laboratory of Membrane Biology at the Peking University School of Life Science (to Yulong Li).
Author information
Authors and Affiliations
Contributions
M.J. and Yulong Li conceived the project. Yuexuan Li, S.Z., T.Q. and M.J. screened the candidate ACh sensors and characterized the sensors in cultured cells and brain slices. J. Zeng and K.T. performed the experiments with transgenic flies. W.P. performed the fiber photometry recordings of ACh signals during the sleep–wake cycle under the supervision of M.X. P.H. performed the fiber photometry recordings of foot-shock-induced ACh signals in the BLA under the supervision of H.L. M.S. and O.K. performed the recordings in VAChT−/− mice under the supervision of L.M.S., V.F.P., T.J.B. and M.A.M.P. S.P. performed the luciferase complementation assay. M.J. and R.W. performed the ACh imaging experiments in the visual cortex using miniature two-photon microscopy under the supervision of L.C. and H.C. J. Zou and S.T. performed imaging in the barrel cortex with the supervision of S.A.H. All authors contributed to the data analysis. M.J. and Yulong Li wrote the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
M.J. and Yulong Li have filed patent applications, the value of which could be affected by this publication.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The engineering process leading to the GRABACh3.0 sensor.
a, Schematic illustration depicting the predicted structure of the generic GRABACh sensor, with the linker region between the receptor (M3R) and cpEGFP magnified at the right and shown in magenta. The crystal structures are from protein database (PDB) archive (PDB ID: 4DAJ for M3R; PDB ID: 3EK4 for cpGFP). b, Site-directed mutagenesis of residues in the N and C termini of the linker region. The numbers indicate amino acid positions in the linker region (the first on N-terminus as N1, and the first on C-terminus as C1). The candidate with the best response is shown in a black circle and is called ACh2.5, with the C4 residue mutated to K; this candidate is used for further engineering steps. c, Left: crystal structure of the cpEGFP moiety in the ACh3.0 sensor; targeted residues for mutagenesis screening are indicated in green and the corresponding amino acid labeled on the structure. Right, the fluorescence response of the indicated mutant candidate sensors is shown on top, with the sequences of the best-performing candidates on the bottom; the relative size of each letter reflects the probability of that amino acid in the sequence. The residues are named by the amino acid followed by the position in cpGFP (the first amino acid in cpGFP as N1). The crystal structures are from protein database (PDB) archive (PDB ID: 3EK4 for cpGFP). d, The fluorescence response of each candidate ACh sensor with combined mutations from the best-performing sites in the linker and cpEGFP. Each point is calculated from the average of >100 cells. e, left, illustration of the ligand binding pocket in M3R, which was mutated from W to A. Right, fluorescence image of HEK293T cells expressing ACh3.0-mut. f, The fluorescence response of ACh3.0 and ACh3.0-mut to indicated concentration of ACh applied (n = 3 wells for each point, with each well averaging >100 cells). Scale bar represents 10 μm. All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM.
Extended Data Fig. 2
Amino acid sequences of different cpGFP-based sensors. The summary of amino acids in linkers and critical residues within cpGFP in different genetically encoded sensors, including GPCR-based sensors and other protein backbone-based sensors.
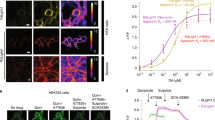
Extended Data Fig. 3 Characterization of ACh2.0 and ACh3.0.
a, The fluorescence response of ACh2.0, ACh3.0, and ACh3.0-mut to 100 μM ACh in HEK293T cells. The fluorescence images are shown on top, and corresponding pseudocolor images representing the signal-to-noise ratio (SNR) are shown at the bottom. Similar results as the representative images were observed for more than 7 cells. Scale bars represent 10 μm. b, The peak fluorescence response (ΔF/F0, left) and SNR (right) of ACh2.0 (black) and ACh3.0 (red) are measured with the indicated concentrations of ACh; n = 8 and 7 cells for ACh2.0 and ACh3.0, respectively. c, Example fluorescence images of ACh3.0 (left) and ACh3.0-mut (right) expressed in cultured rat cortical neurons. Membrane-targeted mScarlet-CAAX is co-expressed and used to confirm expression at the plasma membrane. Similar results as the representative images were observed for more than 5 neurons. Scale bars represent 10 μm in the original image and 5 μm in the magnified images. d, Representative traces (left) and group summary (right) of the fluorescence response of ACh2.0, ACh3.0, and ACh3.0-mut expressed in cultured neurons; where indicated, 100 μM ACh is applied to the cells (n = 4, 5, and 7 neurons for ACh2.0, ACh3.0, and ACh3.0-mut, respectively), p=9.45E-5 between ACh2.0 and ACh3.0; p=6.42E-5 between ACh3.0 and ACh3.0-mut. e, Left, representative traces of the normalized fluorescence change in ACh3.0 (red) and ACh3.0-mut (gray) in response to application of the indicated concentrations of ACh. Note that the ACh-induced fluorescence response in ACh3.0 is blocked by the M3R antagonist tiotropium (Tio, 3 μM). Right, representative trace of the normalized fluorescence change in ACh3.0 in response to indicated compounds. ACh: 100 μM; nicotine (Nic): 50 μM; 5-HT: 1 μM; norepinephrine (NE): 10 μM; dopamine (DA): 20 μM; glutamate (Glu): 10 μM; and Tio: 2 μM. Similar results as the representative images were observed for more than 5 neurons. f, The excitation and emission spectra of ACh3.0 sensor in the absence (light green) and presence of ACh (100 μM, dark green). g, Left, pseudocolor images showing the fluorescence response of ACh3.0 in confocal line scanning mode, with indicated concentrations of ACh applied by bath application. Middle, exemplar fluorescence response trace of ACh3.0 to different concentrations of ACh applied. Right, group data of the ACh3.0 dose-dependent fluorescence response in line scanning mode (from n = 4 coverslips), which is used to estimate the local ACh concentration reaching the cells during kinetics experiments. The steady-state fluorescence response of ACh3.0 to puffed ACh are shown and calibrated based on the curve, with the detail numbers of 10 μM pipette ACh list as an example (pipette short as pip.; Estimated short as Esti.) All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Two-sides Student’s t test performed in (d); ***p<0.001.
Extended Data Fig. 4 The GRABACh3.0 sensor produces negligible downstream signaling.
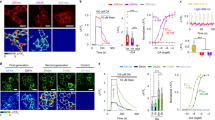
a–c, HEK293T cells expressing either a GFP-tagged M3R construct or ACh3.0 are loaded with the red Ca2+ dye Cal590 (a), and the change in Cal590 fluorescence is measured in response to various concentrations of ACh (b). The Ca2+ influx is calculated as the integration of Cal590 fluorescent signal (ΔF/F0) to ACh application. The group summary data for Ca2+ influx measured in response to 0.1 μM ACh are shown in panel c; n = 21 and 15 cells for GFP-M3R and ACh3.0, respectively, p=1.06E-7. d, Left, cartoon illustrating the experimental design of the luciferase complementation assay, in which cells expressed M3R-SmBit or ACh2.0/3.0-SmBit together with LgBit-mGq. Middle, the luminescence signal measured in non-transfected HEK293T cells (NT), cells expressing ACh2.0/ACh3.0-SmBit, or cells expressing M3R-SmBit in response to application of the indicated concentrations of ACh, normalized to the signal measured in control buffer-treated cells (n = 6 wells for NT; n = 6 wells for M3R; n = 3 wells for ACh2.0; n = 6 wells for ACh3.0, with >100 cells in each well). Right, group summary of the luminescence signal measured in response to 100 μM ACh (n = 6 wells for NT; n = 6 wells for M3R; n = 3 wells for ACh2.0; n = 6 wells for ACh3.0, with >100 cells in each well; p=7.11E-7 between NT and M3R; p=9.95E-7 between M3R and ACh2.0; p=0.003 between ACh2.0 and ACh3.0). e, Similar to (d), except the luminescence signal is measured in HEK293T cells expressing M3R-SmBit or cells expressing both M3R-SmBit and ACh3.0. The group summary at the right shows the luminescence signal in response to 100 μM ACh; n = 5-8 wells per group, with each group averaging >100 cells, p=7.95E-5 between NT and M3R; p=0.33 between M3R and M3R+ACh3.0. f, Schematic cartoon depicting two-photon imaging of transgenic flies in response to odorant stimulation. Ca2+ influx is measured by expressing jRCaMP1a either alone or together with ACh3.0 in the Kenyon cells (KC) of the mushroom body. g, Representative fluorescence traces (left) and group summary (right) of jRCaMP1a fluorescence measured in response to odorant application in flies expressing jRCaMP1a either alone or together with ACh3.0; n = 10 flies per group, p=0.49. All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Scale bar represents 10 μm. Two-sides Student’s t test performed in (c), (d), (e) and (g); ***p<0.001 and n.s., not significant.
Extended Data Fig. 5 Probing endogenous ACh release in mouse brain slices.
a–f, Representative fluorescence traces (a, d) and group summary (b, c, e, f) of the fluorescence change (ΔF/F0 and SNR) in neurons expressing either ACh2.0 or ACh3.0 in response to electrical stimulation in MHb-IPN brain slices. The slices are bathed in either ACSF or 2 μM baclofen (Bac). N=5 slices from 3 mice for ACh2.0, and n = 10 slices from 7 mice for ACh3.0. g, h, The representative fluorescence traces (g) and group data (h) of ACh3.0-expressing neurons in response to 100-Hz electrical stimulation with different stimulation times in MHb-IPN brain slices. The response in either ACSF or 100 μM 4-AP is measured and summarized; n = 5 slices from 5 mice. i, The kinetics of fluorescence response of ACh3.0 to a single pulse (2 ms) of electrical stimulation in the presence of 100 μM 4-AP. The response in three independent experiments are normalized and plotted together in the middle. The group data of on and off response time constants are summarized on the right (n = 3 slices from 3 mice). All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM.
Extended Data Fig. 6 Monitoring in vivo ACh release induced by electrical stimulation in Drosophila.
a, Schematic illustration depicting the experiment in which a transgenic fly expressing ACh3.0 in the KC cells in the mushroom body is placed under a two-photon microscope, and a glass electrode is placed near the mushroom body and used to deliver electrical stimuli. The fly brain is bathed in AHLS containing 100 μM nicotinic acetylcholine receptor blocker mecamylamine (Meca). b, Pseudocolor images (top) and representative traces (bottom) of the fluorescence change in ACh3.0 in response to 2 s of electrical stimulation at the indicated frequencies. Where indicated, the M3R antagonist tiotropium (Tio, 10 μM) is applied to the bath solution. Similar results as the representative images were observed for 8 flies. c, Group summary of the data shown in panel (b); n = 8 flies, p=0.0004. d, ACh3.0 fluorescence is measured before and after a 200-ms electrical stimulation, and the rise and decay phases are fitted with a single-exponential function; the time constants are indicated and summarized on the right; n = 3 flies. All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Two-sides Student’s t test performed in (c); ***p<0.001.
Extended Data Fig. 7 Monitoring endogenous cholinergic signals in BLA of mice in vivo.
a, VAChT immunohistochemistry is performed in coronal mouse sections obtained from a control (VAChT+/+) mouse (left) and a VAChT-/- mouse (right). The insets show magnified views of cholinergic terminals in the basolateral amygdala (BLA). Similar results as the representative images were observed for 6 mice. Scale bars represent 500 μm. b, Diagram summarizing the location of the optic fiber terminals in the BLA of VAChT+/+ (red) and VAChT-/- mice (blue); n = 6 mice per group. This image is modified from the brain map in Allen mouse brain atlas (Allen Mouse Brain Atlas (2004), Allen Institute for Brain Science). c, Pseudocolor image showing the change in ACh3.0 fluorescence measured in the BLA of VAChT+/+ mice (left) and VAChT-/- mice (right) in response to a 1-s foot-shock; ten consecutive trials are shown. d, e, The comparison of ACh2.0 and ACh3.0 fluorescence response in BLA of mice to foot-shock stimuli. The fluorescence signal showing the expression of ACh2.0 and the fiber photometry recording sites are shown in (d). Fluorescence traces and group data of the 1 s foot-shock induced fluorescence response in ACh2.0 (black) and ACh3.0 (red) are shown in (e) (n = 6 mice each for ACh3.0 and ACh2.0, p=0.04). Scale bar: 750 μm. All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Two-sides Student’s t test performed in (e); *, p<0.05.
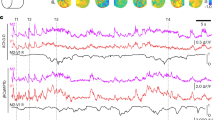
Extended Data Fig. 8 Recording of ACh signal during sleep-wake cycle.
a, The schematic illustration and representative recording data of ACh3.0-mut sensor during the sleep-wake cycle in mice. b, The group data of the fluorescence response of ACh3.0 and ACh3.0-mut sensors in different sleep-wake status (n = 5 mice for ACh3.0 and n = 6 mice for ACh3.0-mut, p=0.0009 in wake; p=0.049 in NREM; p=0.008 in REM). c, The representative recording data of ACh3.0-mut sensor during the sleep-wake cycle in mice. Similar results as the representative images were observed for 6 mice. d, Multiple recording traces of the ACh3.0 sensor during the sleep-wake cycle (from 3 mice, additional to the representative one in Fig. 3g). All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Two-sides Student’s t test performed in (b); *, p<0.05; **, p<0.01; ***, p<0.001.
Extended Data Fig. 9 Imaging of ACh signal in the cortex.
a, Cartoon illustration of the miniature two-photon microscope. b, Group data of ACh3.0 fluorescence in mice recorded while running on a treadmill at indicated speeds (n = 5 mice). c, Representative traces and group summary of ACh3.0 fluorescence measured in mice while performing the running task; where indicated, the mice receive an i.p. injection of saline (black), the nAChR blocker mecamylamine (Meca, 2 mg/kg body weight, blue), or the M3R antagonist scopolamine (Scop, 20 mg/kg body weight, dark yellow); each trace is averaged from 10 trials; n = 5 mice per group, p=0.54 between Saline and Meca; p=0.0002 between Meca and Scop. d, Pseudocolor images showing the ACh3.0 fluorescence response in the S1 in the Hit trial of the whisker-guided object location discrimination task. The left, middle and right image showing the response during baseline, peak in the answer period and after response. Similar results as the representative images were observed for 3 mice. Scale bar: 100 μm. All data are shown as mean value ± SEM, with the error bars or shaded regions indicating SEM. Two-sides Student’s t test performed in (c); *p<0.05, **p<0.01, ***p<0.001, and n.s., not significant.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Jing, M., Li, Y., Zeng, J. et al. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat Methods 17, 1139–1146 (2020). https://doi.org/10.1038/s41592-020-0953-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0953-2
This article is cited by
-
α4 nicotinic receptors on GABAergic neurons mediate a cholinergic analgesic circuit in the substantia nigra pars reticulata
Acta Pharmacologica Sinica (2024)
-
Improved green and red GRAB sensors for monitoring dopaminergic activity in vivo
Nature Methods (2024)
-
Rapid fluctuations in functional connectivity of cortical networks encode spontaneous behavior
Nature Neuroscience (2024)
-
Acetylcholine waves and dopamine release in the striatum
Nature Communications (2023)
-
Sensitive multicolor indicators for monitoring norepinephrine in vivo
Nature Methods (2023)