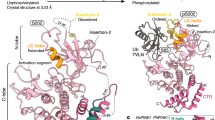
Abstract
Mutations in the ubiquitin ligase parkin are responsible for a familial form of Parkinson’s disease. Parkin and the PINK1 kinase regulate a quality-control system for mitochondria. PINK1 phosphorylates ubiquitin on the outer membrane of damaged mitochondria, thus leading to recruitment and activation of parkin via phosphorylation of its ubiquitin-like (Ubl) domain. Here, we describe the mechanism of parkin activation by phosphorylation. The crystal structure of phosphorylated Bactrocera dorsalis (oriental fruit fly) parkin in complex with phosphorylated ubiquitin and an E2 ubiquitin-conjugating enzyme reveals that the key activating step is movement of the Ubl domain and release of the catalytic RING2 domain. Hydrogen/deuterium exchange and NMR experiments with the various intermediates in the activation pathway confirm and extend the interpretation of the crystal structure to mammalian parkin. Our results rationalize previously unexplained Parkinson’s disease mutations and the presence of internal linkers that allow large domain movements in parkin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
19 July 2018
In the version of this article initially published, RING2 in the schematic to the left in Fig. 1b was mislabeled as RING0. The error has been corrected in the HTML and PDF versions of the article.
References
Martin, I., Dawson, V. L. & Dawson, T. M. Recent advances in the genetics of Parkinson’s disease. Annu. Rev. Genom. Hum. Genet. 12, 301–325 (2011).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Valente, E. M. et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Pickles, S., Vigié, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018).
Trempe, J. F. et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 (2013).
Wauer, T. & Komander, D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 (2013).
Riley, B. E. et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 (2013).
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 (2010).
Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 (2010).
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010).
Vives-Bauza, C. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA 107, 378–383 (2010).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014).
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014).
Sauvé, V. et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 34, 2492–2505 (2015).
Kazlauskaite, A. et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 16, 939–954 (2015).
Wauer, T., Simicek, M., Schubert, A. & Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374 (2015).
Kumar, A. et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 34, 2506–2521 (2015).
Sugiura, A., McLelland, G. L., Fon, E. A. & McBride, H. M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 (2014).
Matheoud, D. et al. Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166, 314–327 (2016).
Wenzel, D. M., Lissounov, A., Brzovic, P. S. & Klevit, R. E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011).
Seirafi, M., Kozlov, G. & Gehring, K. Parkin structure and function. FEBS J. 282, 2076–2088 (2015).
Kumar, A. et al. Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat. Struct. Mol. Biol. 24, 475–483 (2017).
McGinty, R. K., Henrici, R. C. & Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014).
Gladkova, C., Maslen, S., Skehel, J.M. & Komander, D. Mechanism of parkin activation by PINK1. Nature https://doi.org/10.1038/s41586-018-0224-x (2018).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Spratt, D. E. et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat. Commun. 4, 1983 (2013).
Hattori, N. et al. Point mutations (Thr240Arg and Gln311Stop) [correction of Thr240Arg and Ala311Stop] in the Parkin gene. Biochem. Biophys. Res. Commun. 249, 754–758 (1998).
Shimura, H. et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 (2000).
Periquet, M. et al. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain 126, 1271–1278 (2003).
Tang, M. Y. et al. Structure-guided mutagenesis reveals a hierarchical mechanism of Parkin activation. Nat. Commun. 8, 14697 (2017).
Chaugule, V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011).
McLelland, G. L., Soubannier, V., Chen, C. X., McBride, H. M. & Fon, E. A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33, 282–295 (2014).
Abbas, N. et al. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. Hum. Mol. Genet. 8, 567–574 (1999).
Pankratz, N. et al. Parkin dosage mutations have greater pathogenicity in familial PD than simple sequence mutations. Neurology 73, 279–286 (2009).
Safadi, S. S. & Shaw, G. S. A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry 46, 14162–14169 (2007).
Aguirre, J. D., Dunkerley, K. M., Mercier, P. & Shaw, G. S. Structure of phosphorylated UBL domain and insights into PINK1-orchestrated parkin activation. Proc. Natl Acad. Sci. USA 114, 298–303 (2017).
Steffen, J. D., McCauley, M. M. & Pascal, J. M. Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucleic Acids Res 44, 9771–9783 (2016).
Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 112, 6637–6642 (2015).
Okiyoneda, T. et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat. Chem. Biol. 9, 444–454 (2013).
Calles-Garcia, D. et al. Single-particle electron microscopy structure of UDP-glucose:glycoprotein glucosyltransferase suggests a selectivity mechanism for misfolded proteins. J. Biol. Chem. 292, 11499–11507 (2017).
Duda, D. M. et al. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 (2013).
Kelsall, I. R. et al. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 32, 2848–2860 (2013).
Ho, S. R., Lee, Y. J. & Lin, W. C. Regulation of RNF144A E3 ubiquitin ligase activity by self-association through its transmembrane domain. J. Biol. Chem. 290, 23026–23038 (2015).
Smit, J. J. et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844 (2012).
Stieglitz, B., Morris-Davies, A. C., Koliopoulos, M. G., Christodoulou, E. & Rittinger, K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 (2012).
Pao, K. C. et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat. Chem. Biol. 12, 324–331 (2016).
Lazarou, M. et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172 (2013).
Dove, K. K. et al. Structural studies of HHARI/UbcH7~Ub reveal unique E2~Ub conformational restriction by RBR RING1. Structure 25, 890–900.e5 (2017).
Rasool, S. et al. PINK1 autophosphorylation is required for ubiquitin recognition. EMBO Rep. 19, e44981 (2018).
Caulfield, T. R. et al. Phosphorylation by PINK1 releases the UBL domain and initializes the conformational opening of the E3 ubiquitin ligase Parkin. PLOS Comput. Biol. 10, e1003935 (2014).
Ordureau, A. et al. Dynamics of PARKIN-dependent mitochondrial ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Mol. Cell 70, 211–227.e8 (2018).
Geisler, S., Vollmer, S., Golombek, S. & Kahle, P. J. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J. Cell Sci. 127, 3280–3293 (2014).
Dove, K. K. & Klevit, R. E. RING-between-RING E3 ligases: emerging themes amid the variations. J. Mol. Biol. 429, 3363–3375 (2017).
Lechtenberg, B. C. et al. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550 (2016).
Yuan, L., Lv, Z., Atkison, J. H. & Olsen, S. K. Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI. Nat. Commun. 8, 211 (2017).
Martino, L., Brown, N. R., Masino, L., Esposito, D. & Rittinger, K. Determinants of E2-ubiquitin conjugate recognition by RBR E3 ligases. Sci. Rep. 8, 68 (2018).
Cunningham, C. N. et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 17, 160–169 (2015).
Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013).
Kabsch, W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 68, 352–367 (2012).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D. Biol. Crystallogr. 65, 582–601 (2009).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Bai, Y., Milne, J. S., Mayne, L. & Englander, S. W. Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993).
Acknowledgements
We thank J. M. Pascal (Université de Montréal) for the Staphylococcus aureus sortase A construct and M. Seirafi for preparation of proteins for NMR spectroscopy. We thank the Canadian Light Source for access and data collection on the CMCF beamline 08ID-1, and group members for insightful and helpful discussions. We acknowledge support from the Michael J. Fox Foundation and Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Contributions
V.S. designed constructs, froze crystals, performed autoubiquitination assays and prepared HDX samples. V.S. and G.S. performed crystallization and crystal-structure refinement. G.S. performed sortase A reactions. N.S. performed HDX experiments and analysis. G.K., L.M., and N.B. performed NMR experiments. J.-F.T. collected X-ray data, performed phasing and crystal-structure refinement, and analyzed E2-parkin interactions. V.S., G.S., J.-F.T. and K.G. wrote the manuscript. G.L. and K.G. oversaw the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Sequence alignments of parkin orthologs and E2 enzymes.
a, Sequence alignment of parkin orthologs from Bactrocera dorsalis (BD), Rattus norvegicus (RN) and Homo sapiens (HS). b, Sequence alignment of human E2 ubiquitin-conjugating enzymes. The first five E2 enzymes have been reported to function with parkin in vitro or in cells. Blue shading indicates sequence conservation. Residues in the parkin-UbcH7 interface are labeled in green. Residues in the pUbl-RING0 interface are labeled in red. The catalytic cysteines are labeled in yellow
Supplementary Figure 2 Autoubiquitination assay and crystal structures of phosphorylated Bactrocera parkin fused to UbcH7, in the presence of pUb.
a, The autoubiquitination activity of UbcH7-parkin fusion proteins was tested with phosphorylated proteins in the presence of Ub and visualized on SDS-PAGE gel (n = 1). The first two lanes show the activity of UbcH7 and parkin alone as a control. The third to sixth lanes show the activity of the UbcH7-parkin fusions with different length linkers. The linker length between UbcH7 and parkin is 8, 10, 13, 15 amino acids. b, The structure with the REP linker and RING2 present was solved to 4.8 Å using molecular replacement and zinc SAD. Anomalous map was generated from data collected for UbcH7-phosphorylated parkin/pUb crystal at 1.28 Å wavelength. The anomalous dispersion (light grey mesh) confirms the position of zinc atoms (black spheres) of RING0 (green), RING1 (cyan) and IBR (magenta) in the crystal structure of UbcH7-phosphorylated parkin+pUb. Zinc atoms of symmetry-related UbcH7-phosphorylated parkin+pUb molecules within the crystal are displayed in light orange. No unassigned anomalous density was observed. c, The structure without the disordered elements was solved to 3.8 Å using molecular replacement using 4.8 Å structure as the model. The all-atom root mean square deviation between the two structures is 0.2 Å
Supplementary Figure 3 Steric interactions regulate parkin activity.
a, Overlay of activated Bactrocera parkin structure (colored) with the RING2 modeled from the inactive parkin (grey, PDB 4K95). The C-terminal helix of RING2 occupies the same position as pUbl residues preceding the phosphoserine residue (pSer94). b, Overlay of UbcH7 with the Ubl domain from inactive parkin. The Ubl domain prevents UbcH7 residues Glu60 and Phe63 from binding to residues Gln301, Tyr302, Ser305, and Arg 306 of helix 1 of the parkin RING1 domain. c, Overlay of UbcH7 with the REP linker from inactive parkin. The REP residues preceding the helix block contact of UbcH7 residues Lys96, Pro97, and Ala98 with Bactrocera parkin residues Ala326 and Leu274
Supplementary Figure 4 Electrostatic interactions between UbcH7 and parkin RING1.
a, Structure of Bactrocera parkin bound to UbcH7. An electrostatic potential map was calculated for the isolated RING1 domain. The side-chains of Arg5 and Arg6 in UbcH7 interact with a negatively charged surface in RING1. b, Model of rat Parkin (PDB 4ZYN) bound to UbcH7 with an electrostatic potential map computed for the isolated RING1 domain (REP and Ubl removed)
Supplementary Figure 5 Phosphorylation of parkin mutants for ubiquitin assays.
a, For the autoubiquitination assay, the mutants and WT were phosphorylated and verified on a 7.5% phos-tag SDS-PAGE (n = 1 assay). b, For the complementation assay, the mutants and WT were phosphorylated and verified on a 7.5 % phos-tag SDS-PAGE (n = 1 assay). c, Intact mass spectrometry of the phosphorylated proteins, collected on a Impact II ESI-QTOF (n = 1 assay). The predicted average mass is displayed on the left.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5
Supplementary Dataset 1
Uncropped NMR spectra and gel from Figure 3
Supplementary Dataset 2
Graphic presentation of HDX-MS data from Figure 4
Supplementary Dataset 3
Excel data of HDX-MS data from Figure 4
Rights and permissions
About this article
Cite this article
Sauvé, V., Sung, G., Soya, N. et al. Mechanism of parkin activation by phosphorylation. Nat Struct Mol Biol 25, 623–630 (2018). https://doi.org/10.1038/s41594-018-0088-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0088-7
This article is cited by
-
Optineurin provides a mitophagy contact site for TBK1 activation
The EMBO Journal (2024)
-
The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family
Nature Communications (2023)
-
The mechanisms and roles of selective autophagy in mammals
Nature Reviews Molecular Cell Biology (2023)
-
Discovery of Small Molecule PARKIN Activator from Antipsychotic/Anti-neuropsychiatric Drugs as Therapeutics for PD: an In Silico Repurposing Approach
Applied Biochemistry and Biotechnology (2023)
-
Activation mechanism of PINK1
Nature (2022)