Abstract
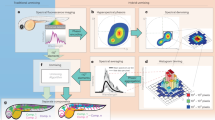
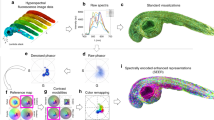
Fluorescence lifetime imaging microscopy (FLIM) is used in diverse disciplines, including biology, chemistry and biophysics, but its use has been limited by the complexity of the data analysis. The phasor approach to FLIM has the potential to markedly reduce this complexity and at the same time provide a powerful visualization of the data content. Phasor plots for fluorescence lifetime analysis were originally developed as a graphical representation of excited-state fluorescence lifetimes for in vitro systems. The method's simple mathematics and specific rules avoid errors and confusion common in the study of complex and heterogeneous fluorescence. In the case of FLIM, the phasor approach has become a powerful method for simple and fit-free analyses of the information contained in the many thousands of pixels constituting an image. At present, the phasor plot is used not only for FLIM, but also for hyperspectral imaging, wherein phasors provide an unprecedented understanding of heterogeneous fluorescence. Undoubtedly, phasor plots will be increasingly important in the future analysis and understanding of FLIM and hyperspectral confocal imaging. This protocol presents the principle of the method and guides users through one of the popular interfaces for FLIM phasor analysis, namely, the SimFCS software. Implementation of the analysis takes only minutes to complete for a dataset containing hundreds of files.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
References
Stringari, C. et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA 108, 13582–13587 (2011).
Hinde, E., Digman, M. A., Hahn, K. M. & Gratton, E. Millisecond spatiotemporal dynamics of FRET biosensors by the pair correlation function and the phasor approach to FLIM. Proc. Natl. Acad. Sci. USA 110, 135–140 (2013).
Hinde, E., Digman, M. A., Welch, C., Hahn, K. M. & Gratton, E. Biosensor Forster resonance energy transfer detection by the phasor approach to fluorescence lifetime imaging microscopy. Microsc. Res. Tech. 75, 271–281 (2012).
Battisti, A. et al. Intracellular pH measurements made simple by fluorescent protein probes and the phasor approach to fluorescence lifetime imaging. Chem. Commun. 48, 5127 (2012).
Celli, A. et al. The epidermal Ca2+ gradient: measurement using the phasor representation of fluorescent lifetime imaging. Biophys. J. 98, 911–921 (2010).
Ma, N., Digman, M. A., Malacrida, L. & Gratton, E. Measurements of absolute concentrations of NADH in cells using the phasor FLIM method. Biomed. Opt. Express 7, 2441–2452 (2016).
Harvey, B. J. & Levitus, M. Nucleobase-specific enhancement of Cy3 fluorescence. J. Fluoresc. 19, 443–448 (2009).
Malacrida, L., Jameson, D. M. & Gratton, E. A multidimensional phasor approach reveals LAURDAN photophysics in NIH-3T3 cell membranes. Sci. Rep. 7, 9215 (2017).
Datta, R., Heylman, C., George, S. C. & Gratton, E. Label-free imaging of metabolism and oxidative stress in human induced pluripotent stem cell-derived cardiomyocytes. Biomed. Opt. Express 7, 1690 (2016).
Datta, R., Alfonso-García, A., Cinco, R. & Gratton, E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Sci. Rep. 5, 9848 (2015).
Ranjit, S. R. et al. Measuring the effect of a western diet on liver tissue architecture by FLIM autofluorescence and harmonic generation microscopy. Biomed. Opt. Express 8, 371–378 (2017).
Weber, G. Resolution of the fluorescence lifetimes in a heterogeneous system by phase and modulation measurements. J. Phys. Chem. 85, 949–953 (1981).
Jameson, D. M., Gratton, E. & Hall, R. D. The measurement and analysis of heterogeneous emissions by multifrequency phase and modulation fluorometry. Appl. Spectrosc. Rev. 20, 55–106 (1984).
Verveer, P. J., Squire, A. & Bastiaens, P. I. Global analysis of fluorescence lifetime imaging microscopy data. Biophys. J. 78, 2127–2137 (2000).
Pelet, S., Previte, M. J. R., Laiho, L. H. & So, P. T. C. A fast global fitting algorithm for fluorescent lifetime imaging microscopy based on image segmentation. Biophys. J. 87, 2807–2817 (2004).
Jameson, D. M. Introduction to Fluorescence (CRC Press, Boca Raton, FL, 2014).
O’Connor, D. & Phillps, D. Time-Correlated Single Photon Counting (Academic Press, New York, 1984).
Gratton, E. in Perspectives on Fluorescence: A Tribute to Gregorio Weber (ed. Jameson, D. M.) 67–80 (Springer International Publishing, Switzerland, 2016).
Arnesano, C., Santoro, Y. & Gratton, E. Digital parallel frequency-domain spectroscopy for tissue imaging. J. Biomed. Opt. 17, 0960141 (2012).
Colyer, R. A., Lee, C. & Gratton, E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc. Res. Tech. 71, 201–213 (2008).
Digman, M. A. & Gratton, E. in Fluorescence Lifetime Spectroscopy and Imaging: Principles and Applications in Biomedical Diagnostics (eds. Marcu, L., French, P. M. W. & Elson, D. S.) 235–248 (CRC Press, Boca Raton, FL, 2014)
Kristoffersen, A. S., Erga, S. R., Hamre, B. & Frette, Ø. Testing fluorescence lifetime standards using two-photon excitation and time-domain instrumentation: Rhodamine B, coumarin 6 and lucifer yellow. J. Fluoresc. 24, 1015–1024 (2014).
Chance, B. Mitochondrial NADH redox state, monitoring discovery and deployment in tissue. Methods Enzymol. 385, 361–370 (2004).
Mayevsky, A. & Chance, B. Oxidation-reduction states of NADH in vivo: from animals to clinical use. Mitochondrion 7, 330–339 (2007).
Zhang, Z. et al. Metabolic imaging of tumors using intrinsic and extrinsic fluorescent markers. Biosens. Bioelectron. 20, 643–650 (2004).
Bird, D. K. et al. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 65, 8766–8773 (2005).
Skala, M. C. et al. In vivo multiphoton microscopy of {NADH} and {FAD} redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA 104, 19494–19499 (2007).
Chance, B., Nioka, S., Warren, W. & Yurtsever, G. in Oxygen Transport to Tissue XXVI (eds. Okunieff, P., Williams, J. & Chen, Y.) 231–242 (Springer, USA, 2005).
Nakashima, N., Yoshihara, K., Tanaka, F. & Yagi, K. Picosecond fluorescence lifetime of the coenzyme of D-amino acid oxidase*. J. Biol. Chem. 255, 5261–5263 (1980).
Stringari, C., Nourse, J. L., Flanagan, L. A. & Gratton, E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS ONE 7, e48014 (2012).
Aguilar-Arnal, L. et al. Spatial dynamics of SIRT1 and the subnuclear distribution of NADH species. Proc. Natl. Acad. Sci. USA 113, 12715–12720 (2016).
Valeur, B. in Molecular Fluorescence: Principles and Applications 247–272 (Wiley-VCH, Weinheim, Germany, 2001).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005).
DiPilato, L. M. & Zhang, J. Fluorescent protein-based biosensors: resolving spatiotemporal dynamics of signaling. Curr. Opin. Chem. Biol. 14, 37–42 (2010).
Sun, Y., Day, R. N. & Periasamy, A. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat. Protoc. 6, 1324–1340 (2011).
Day, R. N., Tao, W. & Dunn, K. W. A simple approach for measuring FRET in fluorescent biosensors using two-photon microscopy. Nat. Protoc. 11, 2066–2080 (2016).
Digman, M. A., Caiolfa, V. R., Zamai, M. & Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 (2008).
Kolb, D. A. & Weber, G. Quantitative demonstration of the reciprocity of ligand effects in the ternary complex of chicken heart lactate dehydrogenase with nicotinamide adenine dinucleotide and oxalate. Biochemistry 14, 4471–4476 (1975).
Jameson, D. M., Thomas, V. & Zhou, D.-M. Time-resolved fluorescence studies on NADH bound to mitochondrial malate dehydrogenase. Biochim. Biophys. Acta 994, 187–190 (1989).
Stringari, C., Sierra, R., Donovan, P. J. & Gratton, E. Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy. J. Biomed. Opt. 17, 046012 (2012).
Scott, T. G., Spencer, R. D., Leonard, N. J. & Weber, G. Emission properties of NADH. Studies of fluorescence lifetimes and quantum efficiencies of NADH, AcPyADH, and simplified synthetic models. J. Am. Chem. Soc. 7302, 687–695 (1969).
Hirshfield, K. M., Toptygin, D., Packard, B. S. & Brand, L. Dynamic fluorescence measurements of two-state systems: applications to calcium-chelating probes. Anal. Biochem. 209, 209–218 (1993).
Pineda Rodó, A., Váchová, L. & Palková, Z. In vivo determination of organellar pH using a universal wavelength-based confocal microscopy approach. PLoS ONE 7, e33229 (2012).
Okabe, K. et al. Intracellular temperature mapping with a fluorescent polymeric thermometer and fluorescence lifetime imaging microscopy. Nat. Commun. 3, 705 (2012).
Ma, Y. et al. Phosphorescent soft salt for ratiometric and lifetime imaging of intracellular pH variations. Chem. Sci. 7, 3338–3346 (2016).
Sherin, P. S. et al. Visualising the membrane viscosity of porcine eye lens cells using molecular rotors. Chem. Sci. 8, 3523–3528 (2017).
Van Den Zegel, M., Boens, N., Daems, D. & De Schryver, F. C. Possibilities and limitations of the time-correlated single photon counting technique: a comparative study of correction methods for the wavelength dependence of the instrument response function. Chem. Phys. 101, 311–335 (1986).
Luchowski, R. et al. Fluorescence instrument response standards in two-photon time-resolved spectroscopy. Appl. Spectrosc. 64, 918–922 (2010).
Talbot, C. B. et al. Application of ultrafast gold luminescence to measuring the instrument response function for multispectral multiphoton fluorescence lifetime imaging. Opt. Express 19, 13848–13861 (2011).
Becker, W. Recording the instrument response function of a multiphoton FLIM system. Becker & Hickl: Application Notes http://www.becker-hickl.com/pdf/irf-mp04.pdf (2008).
Wahl, M. Time-correlated single photon counting. PicoQuant: Technical Notes https://www.picoquant.com/images/uploads/page/files/7253/technote_tcspc.pdf (2014).
Becker, W. The bh TCSPC Handbook (Becker & Hickl, Berlin, Germany, 2014).
Štefl, M., James, N. G., Ross, J. A. & Jameson, D. M. Applications of phasors to in vitro time-resolved fluorescence measurements. Anal. Biochem. 410, 62–69 (2011).
Reinhart, G. D., Marzola, P., Jameson, D. M. & Gratton, E. A method for on-line background subtraction in frequency domain fluorometry. J. Fluoresc. 1, 153–162 (1991).
Acknowledgements
This work was supported in part by NIH grants NIH P41-GM103540 and NIH P50-GM076516. L.M. was supported as a full-time professor at the Universidad de la República-Uruguay.
Author information
Authors and Affiliations
Contributions
S.R. and L.M. wrote the manuscript and prepared the figures. E.G. and D.M.J. wrote the introduction and corrected all preliminary documents. All data in the article were collected by S.R. and L.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
1. Stringari, C. et al. Proc. Natl. Acad. Sci. USA 108, 13582–13587 (2011) https://doi.org/10.1073/pnas.1108161108.
2. Hinde, E., Digman, M. A., Welch, C., Hahn, K. M. & Gratton, E. Microsc. Res. Tech. 75, 271–281 (2012) https://doi.org/10.1002/jemt.21054.
3. Digman, M. A., Caiolfa, V. R., Zamai, M. & Gratton, E. Biophys. J. 94, L14–L16 (2008) https://doi.org/10.1529/biophysj.107.120154.
4. Ma, N., Digman, M. A., Malacrida, L. & Gratton, E. Biomed. Opt. Express 7, 441–2452 (2016) https://doi.org/10.1364/BOE.7.002441.
Supplementary information
Combined Supplementary Information
Supplementary Figs. 1–18, Supplementary Methods and Supplementary Notes 1–6
Rights and permissions
About this article
Cite this article
Ranjit, S., Malacrida, L., Jameson, D.M. et al. Fit-free analysis of fluorescence lifetime imaging data using the phasor approach. Nat Protoc 13, 1979–2004 (2018). https://doi.org/10.1038/s41596-018-0026-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0026-5
This article is cited by
-
More than double the fun with two-photon excitation microscopy
Communications Biology (2024)
-
Histone FRET reports the spatial heterogeneity in nanoscale chromatin architecture that is imparted by the epigenetic landscape at the level of single foci in an intact cell nucleus
Chromosoma (2024)
-
Visualising varnish removal for conservation of paintings by fluorescence lifetime imaging (FLIM)
Heritage Science (2023)
-
Unveiling nanoscale optical signatures of cytokine-induced β-cell dysfunction
Scientific Reports (2023)
-
Biomolecular condensates modulate membrane lipid packing and hydration
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.