Abstract
Diverse pathogenic fungi secrete extracellular vesicles (EV) that contain macromolecules, including virulence factors that can modulate the host immune response. We recently demonstrated that the binding of monoclonal antibodies (mAb) modulates how Histoplasma capsulatum load and releases its extracellular vesicles (EV). In the present paper, we addressed a concentration-dependent impact on the fungus’ EV loading and release with different mAb, as well as the pathophysiological role of these EV during the host-pathogen interaction. We found that the mAbs differentially regulate EV content in concentration-dependent and independent manners. Enzymatic assays demonstrated that laccase activity in EV from H. capsulatum opsonized with 6B7 was reduced, but urease activity was not altered. The uptake of H. capsulatum by macrophages pre-treated with EV, presented an antibody concentration-dependent phenotype. The intracellular killing of yeast cells was potently inhibited in macrophages pre-treated with EV from 7B6 (non-protective) mAb-opsonized H. capsulatum and this inhibition was associated with a decrease in the reactive-oxygen species generated by these macrophages. In summary, our findings show that opsonization quantitatively and qualitatively modifies H. capsulatum EV load and secretion leading to distinct effects on the host’s immune effector mechanisms, supporting the hypothesis that EV sorting and secretion are dynamic mechanisms for a fine-tuned response by fungal cells.
Similar content being viewed by others
Introduction
Histoplasmosis is a systemic mycosis caused by Histoplasma capsulatum, a thermodimorphic fungus that primarily exists in a filamentous form at temperatures between 25 and 28 °C and undergoes morphogenesis to a yeast at 37 °C. Infection occurs after inhalation of micropropagules, which leads to a primary pulmonary disorder. Although infection is common, the majority of individuals have only mild or moderate symptoms. However, the fungus can disseminate throughout the body causing severe disease and death1,2,3.
The onset of the host protective response involves the rapid phagocytosis of fungal cells by neutrophils and macrophages, and the subsequent activation of a Th1 response4,5. Phagocytosis can occur through different molecular mechanisms, though H. capsulatum mainly engages macrophages through Fcγ receptors (IgG-opsonized H. capsulatum) and by complement receptor 3 (CR3). CR3 (CD11b/CD18) can recognize the heat shock protein 60 (Hsp60) on the surface of H. capsulatum and promote the internalization of the fungus6,7. Through diverse mechanisms, including regulation of vacuolar ATPases8 and iron acquisition9,10, H. capsulatum evades the host immune system and multiplies. Our group previously demonstrated that opsonization of H. capsulatum with monoclonal antibodies (mAbs) against Hsp60 can change the outcome of histoplasmosis in vitro and in vivo. In addition, administration of 6B7 mAb (IgG1) was protective while 7B6 mAb (IgG2b) failed to protect in macrophage studies or in murine infection models, despite the fact that both mAbs recognize the same epitope in Hsp6011. Moreover, we recently established that treatment of H. capsulatum with these mAbs significantly changed the characteristics and contents of extracellular vesicles (EV) produced by the yeast cells12. Fungal EV are spherical, bilayered compartments with diameters ranging from 20 to 500 nm that can carry lipids, carbohydrates, proteins, pigments and nucleic acids13,14, many of which are constituents of the fungal cell wall and diverse others are associated with stress response and pathogenesis, such as urease, phosphatase, catalase and laccase15,16,17. The fact that antibody directly affects EV biology extends the physiological actions of these molecules.
Given that mAb modifies H. capsulatum pathogenesis and EV content, we decided to delve deeper into associations between these findings. First, we examined if antibody-mediated changes on EV secretion and protein sorting are concentration dependent. Additionally, we assessed whether the EV produced in the presence of mAb differentially impacted host cell function by determining whether macrophages exposed to the EV had alterations in the capacity to phagocytosis and kill H. capsulatum yeast cells.
Results
Protein and sterol quantification in EV derived from Histoplasma capsulatum cells
Protein and sterol content were evaluated in the EV’s suspension. For practical purposes, EV secreted by H. capsulatum opsonized with 6B7 and 7B6 will henceforward be referred as 6B7.6-EV and 7B6.6-EV (6 μg/mL of mAb) or 6B7.20-EV and 7B6.20-EV (20 μg/mL of mAb) respectively. Similar to what we found with 6 µg/mL of mAb12, 6B7.20-EV and 7B6.20-EV had an increase in the protein/sterol ratio, when compared to EV from untreated yeast cells (Fig. 1A). In addition, the 7B6.20-EV presented higher protein/sterol ratios compared to either EV from untreated yeast or 6B7.20-EV (Fig. 1B). Interestingly, an increase of sterol content was observed only in the 7B6.20-EV group. EV from H. capsulatum are divided in two populations, a smaller one with 50 nm and a bigger one with 200 nm. Both populations of 6B7.20-EV are bigger than control EV, while just the smaller population of 7B6.20-EV are bigger than the control EV (Supplementary Fig. S1).
Protein and sterol quantification in EV from H. capsulatum. Yeast cells were incubated with or without 6 and 20 μg/mL of 6B7 and 7B6 mAb. The EV protein content was determined by BCA assay (A). EV sterol quantification was performed using Amplex reagent kit (B). Graphs represent means and standard deviation from at least two independent EV isolations and all the analyses were performed in duplicate. *p ≤ 0.05, compared to the untreated control group. #p ≤ 0.05, compared to the groups 6B7 and 7B6 at 6 µg/mL. ϕp ≤0.05, compared to 6B7 mAb treatment at 20 µg/mL.
Enzymatic activity of fungal virulence factors in EV after treatments with mAb
The activities of urease and laccase were evaluated in the suspensions of EV by the addition of chromogenic substrates specific to each enzyme. There was no difference in urease activity among the groups, even when the antibody concentration increased from 6 µg/mL to 20 µg/mL (Fig. 2A). In contrast, laccase activity was significantly reduced in 6B6.6-EV and 6B6.20-EV, but unaltered in 7B6.6-EV and 7B6.20-EV (Fig. 2B).
Enzymatic activity of EV virulence factors resulting from fungal opsonization with mAb. Yeast cells were incubated with or without 6 and 20 μg/mL of 6B7 or 7B6 mAb. After EV isolation, urease (A) and laccase (B) activities were measured as described in Methods. Graphs represent means and standard deviation of at least two independent experiments and all the analyses were performed in duplicate. *p ≤ 0.05 compared to control EV.
Proteomic analysis of EV from H. capsulatum treated with either 6B7 or 7B6 mAb
H. capsulatum yeasts were treated with 20 µg/mL of mAbs, 6B7 or 7B6, and released vesicles were purified and analyzed by proteomic analysis. We further compared this data with our previously published data on EV derived from yeasts treated with 6 µg/mL of 6B7 and 7B6 mAbs12. Combining the 6 and 20 µg/mL conditions, a total of 936 proteins were detected above the limit of quantifications and use for comparative analysis (Supplemental Table S1). Combining both antibody concentration treatments, 170 and 131 proteins were differentially abundant in response to 7B6 and 6B7 mAbs, respectively, with 250 proteins differentially represented in EV resulting from H. capsulatum cultures previously incubated with mAbs (Table 1). To better understand possible functions of the EV proteins affected by the antibody treatment, we performed an annotation based on the Kyoto Encyclopedia of Genes and Genome (KEGG), followed by a function-enrichment analysis. The most affected pathways were biosynthesis of amino acids, glycolysis/ gluconeogenesis, fatty acid degradation, and vitamin B6 metabolism (Table 1). Pathways, such as biosynthesis of amino acids, ribosomes, spliceosome, protein processing in the endoplasmic reticulum might have a direct effect on the protein composition of the yeast and consequently in the EV cargo.
To have a view of the general trends of the antibody-induced changes in the EV composition, we clustered the proteins based on their abundance profiles using the K-mean method and plotted heat maps of each cluster. Three very distinct clusters were obtained. The first cluster had proteins whose abundances were changing in response to the antibody concentration, increased in abundances with 6 µg/mL of antibody treatment, but decreased in the higher concentration. Pathways, such as ribosomes and biosynthesis of amino acids, were enriched in this cluster (Fig. 3). The second cluster was enriched in proteins that were upregulated with the treatment, independent of the antibody concentration, and was represented by functions, such as fatty acid degradation, oxidative phosphorylation and phagosome (Fig. 3). The last cluster had proteins that were downregulated with 6 µg/mL of antibody treatment, but were less affected with the higher concentration. This cluster was particularly enriched with proteins of the central carbon metabolism (Fig. 3).
Heatmap of proteins differentially abundant in response to treatment with 6 or 20 µg/mL of 6B7 and 7B6 mAb against Hsp60. The heatmap was generated using Multiexperiment Viewer and clustered by K-means based on the abundance profiles. The significantly enriched functions (p ≤ 0.05 by Fisher’s exact test, 2 folds enrichment compared to the genome background) are listed for each cluster. Gray spots represent hits below the limit of quantification.
We next investigated the differences in EV composition after 6B7 and 7B6 mAb treatments. A total of 72 proteins were differentially abundant comparing both antibody treatments and were overrepresented in pathways such as central carbon metabolism, protein synthesis (Ribosome and Protein processing in endoplasmic reticulum) and the phagosome. The regulation of some proteins was dependent on the antibody concentration while others were independent (Fig. 4).
Heatmap of proteins differentially abundant in EV in response to treatment of H. capsulatum with 6B7 and 7B6 mAb against Hsp60. The heatmap was generated using Multiexperiment Viewer and clustered by hierarchical clustering based on the abundance profiles. The significantly enriched functions (p ≤ 0.05 by Fisher’s exact test, 2 folds enrichment compared to the genome background) are listed for each cluster. The gray spot represents hits below the limit of quantification.
Effect of extracellular vesicles on macrophages
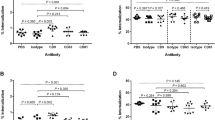
We investigated a possible effect of distinct EV on the effector functions of bone marrow-derived macrophages (BMDM). BMDM were treated for 1 hour with EV from control or opsonized H. capsulatum prior to the in vitro challenge with H. capsulatum yeast cells (Fig. 5A). The inhibition of actin microfilaments dynamics with cytochalasin B largely abrogated phagocytosis. Treatment of murine BMDM with control EV inhibited H. capsulatum phagocytosis by 35% compared to non-treated BMDM. The same effect was observed when BMDM were treated with 6B7.20-EV or 7B6.20-EV. Interestingly 6B7.6-EV and 7B6.6-EV promoted an even stronger inhibition of phagocytosis compared to control EV-treated BMDM.
Phagocytosis and killing of H. capsulatum by EV-treated macrophages. BMDMs were treated for 1 hour with EV from H. capsulatum opsonized or not with 6 or 20 μg/mL of 6B7 or 7B6 mAb prior to the in vitro challenge with H. capsulatum-GFP (m.o.i. 1:5). After 1 hour, phagocytosis was analyzed by flow cytometry (A). To evaluate intracellular killing, after the phagocytosis time, and removal of extracellular yeast cells, macrophages were incubated for additional 2 hours and then lysed. Lysates were plated onto BHI-agar plates and colonies were counted. Graphs show the mean and standard errors from 3 independent experiments performed in triplicates. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 compared to untreated cells, and +p < 0.05 compared to control EV, by Student’s t-test; Cytochalasin B (10 μM).
This data suggest that EV released by H. capsulatum can inhibit the phagocytosis of H. capsulatum by BMDM, and EV secreted by H. capsulatum opsonized with the lower concentration either 6B7 or 7B6 mAb are even more potent than control EV in inhibiting the phagocytosis of H. capsulatum yeast cells.
The intracellular killing of H. capsulatum by BMDM treated with control EV, 6B7.20-EV or 7B6.20-EV was inhibited when compared to untreated BMDM (Fig. 5B). Interestingly, the treatment with 6B7.6-EV did not promote any significant effect on the intracellular killing of H. capsulatum by BMDM, even though there was a notable trend towards the inhibition. Nevertheless, the treatment with 7B6.6-EV led to an inhibition of the yeast intracellular killing by BMDM even stronger than the control EV. These data suggest that the opsonization of H. capsulatum with the non-protective mAb (7B6) induces the release of EV that promote a more potent inhibition on the intracellular killing of the yeast by BMDM than the control EV.
The same assays have slightly different results when performed with human macrophages (THP-1-derived macrophages). The only treatment that inhibited phagocytosis was 7B6.20-EV, but the intracellular killing was inhibited by control EV as well as 6B7.20-EV and 7B6.20-EV. As seen for murine macrophages, the intracellular killing of H. capsulatum yeast cells by human macrophages was strongly inhibited after treatment with 7B6.6-EV, supporting the non-protective effect of this mAb (Supplementary Figure S2).
To further address the intracellular killing inhibition of H. capsulatum by macrophages, we evaluated the reactive-oxygen species (ROS) production by macrophages that were treated with EV and then challenged with H. capsulatum yeast cells (Fig. 6).
Reactive oxygen species generation by EV-treated macrophages. BMDMs were loaded with H2DCFDA and then treated or not with EV from H. capsulatum opsonized or not with 6 or 20 μg/mL of 6B7 or 7B6 mAb, prior to the incubation with H. capsulatum yeast cells. ROS generation was measured in a microplate reader 2 hours after the incubation of the yeast cells. Graphs show the mean and standard errors from 2 independent experiments performed in quadruples. ****p < 0.0001 compared to macrophages without EV treatment. +p < 0.05; ++p < 0.01 and ++++p < 0.0001 compared to macrophages treated with control EV, by One-way ANOVA followed by Tukey’s multiple comparisons test.
Curiously, the treatment of BMDM with control EV promoted a (non-significant) trend in increasing the ROS generation when compared to untreated cells. However, the treatment with either 7B6.6-EV or 7B6.20-EV markedly inhibited ROS production by BMDM when compared to both untreated and control EV-treated BMDM. 6B7.6-EV and 6B7.20-EV did not promote any change in ROS generation by BMDM when compared to untreated cells, but a slight difference was noted compared to control EV-treated BMDM.
Discussion
EV are secreted by cells from a remarkably evolutionary divergent range of organisms, including bacteria, protozoan, fungi and mammalian cells13. EV secretion by pathogenic organisms facilitates their survival and promotes disease through the delivery of diverse simple and complex molecules (toxins, enzymes, etc.) that subvert different hosts’ immune systems18,19. Given that EV from pathogenic organisms are loaded with immunogens and inflammatory activators that can efficiently trigger immune responses, EV have even been harnessed for vaccine delivery and animal models show that EV can confer protection to a host18,20,21,22. Despite advances in our knowledge of EV biology by diverse microbes, the mechanisms involved with EV biogenesis and cargo sorting as well as the roles of EV in fungal pathogenesis are poorly understood. By using mAbs against Hsp60, a protein exposed at the surface of H. capsulatum yeast cells, we recently described that mAbs have the capacity to alter the characteristics and cargo of EV12. In the current work we quantitatively examined alterations in protein abundance and determined whether host effector functions were affected by EV derived from H. capsulatum treated or not with 6 or 20 μg/mL of either protective (6B7) or non-protective (7B6) mAb. The amount of protein in EV samples increased when H. capsulatum was treated with either mAb at 6 or 20 μg/mL, although ergosterol concentration did not change even when the antibody concentration increased from 6 μg/mL to 20 μg/mL. The exception was for 7B6.20-EV compared to control EV. Fundamentally, mAb appears to function as a stress signal that leads the fungal cell to export more proteins through enhanced loading of EV.
The biological functions of 6B7 and 7B6 mAbs are dichotomous, with 6B7 mAb protecting the host against H. capsulatum infection, while the 7B6 mAb is irrelevant or even deleterious11. The hypothesis that mAbs act as a stressor for H. capsulatum is sustained by the assessment of the enzymatic activity of known virulence factors in isolated EV. Laccase is a well-known promoter of virulence in fungi23 and it is present in H. capsulatum yeast cells24. As we previously reported for EV collected from H. capsulatum treated with 6 μg/mL of 6B7 or 7B6 mAb12, laccase activity significantly decreased after treatment with 20 μg/mL of the protective 6B7 antibody, but not with 7B6 mAb. However, the modulation of enzymatic activity found in EV was not global, as urease activity, a virulence factor that facilitate fungal survival and proliferation in the host tissue, was not altered by the incubation of the fungus with mAb25,26.
The proteomic data shows that the most impacted pathways after treatment with both subtypes of antibodies were biosynthesis of amino acid, glycolysis/gluconeogenesis and fatty acid degradation. The amount of heat shock protein (C0NVB8) and heat-shock protein 60-like protein (C0P0B3) found in EV consistently decreased among all mAb treatments when compared to control, suggesting a possible feedback mechanism triggered by mAb binding to hsp-60. The presence of chitinase (C0NW75) in EV was also equally downregulated among all treatments. This data suggests that the yeast’s cell wall could be somehow impacted by the mAb binding, as we can also infer by analyzing the presence of the cell wall synthesis protein (C0ND43), which is increased after treatment with 6B7, but decreased after 7B6 mAb. The extracellular cell wall glucanase Crf1 (C0NSG6) was upregulated inside EV only after treatment with 6B7 mAb. This finding is very interesting, as this protein has proven to be highly immunogenic in Candida albicans and Aspergillus fumigatus and capable of eliciting a Th1 response in these models27.
In response to antibody treatments some proteins increase in abundance with 6 μg/mL of mAb, but decrease after treatment with 20 μg/mL. These differences suggest that the mAbs activate certain pathways in the yeast when in a lower concentration, but higher mAb concentrations dampen some of these pathways, by a possible desensitization of the receptor(s) involved, due to the presence of high amounts of ligand. In addition, after mAb treatment, proteins can increase in abundance independent of the antibody concentration or decrease in abundance with 6 μg/mL of antibody, but less affected with a higher concentration.
Phagocytosis of H. capsulatum is an important defense mechanism displayed by host phagocytes in order to control the infection28. However, during the co-evolution of pathogen and mammalian phagocytes29 as well as pathogen interactions with environmental predators30, H. capsulatum has developed a remarkable ability to live inside macrophages. Thus, the signaling pathways activated during the contact between macrophages and H. capsulatum are crucial for determining the intracellular fate of the pathogen. Interestingly, we observed that macrophages treated with EV were less able to kill fungal cells, mainly phagocytes treated with 7B6-EV. The treatment with 6B7 mAb decreased the amount of glutathione peroxidase (C0NI23) in EV. Glutathione peroxidase participates in the metabolism of glutathione, which is important for the detoxification of peroxides. Thus, the lower abundance of glutathione peroxidase in 6B7-EV could help the yeast cells to deal with peroxides generated by phagocytes. However, the intracellular killing of H. capsulatum by BMDM was slightly inhibited after the treatment with 6B7-EV but this inhibition was much stronger when BMDM were treated with 7B6-EV. This result suggests that, the scavenging of reactive-oxygen species (ROS) as well as a direct inhibition on macrophage’s NADPH oxidase and/or NO synthase are possible explanations to answer how EV increase the intracellular yeast survival.
In addition to its role as a chromatin protein, histone 2B is a cell surface antigen on H. capsulatum that is associated with binding to host cells and it also modulates the intracellular fate of the fungus31,32. The decrease in abundance of histone 2B after opsonization with 6B7 mAb may decrease the signal for fungal cell proliferation leading to a reduction of its intracellular replication31. These differences in the abundance profiles corroborate the idea that each mAb induces important changes in fungal cells that subsequently affect fungal-host interactions.
The abundance of coronin (C0NA44) and cofilin (C0P0B4), both actin cytoskeleton adaptor molecules, is consistently regulated by the yeast opsonization with either 6B7 or 7B6 mAb, while proteins such as actin (P53455), tubulin (C0NKB3), vacuolar sorting-associated protein (C0NT96) and Svf1 family protein (C0NFT1) are increased in abundance by 7B6 relative to 6B7 treatments. Differences in the abundance of these proteins suggest intense movement and high protein processing in the cytoplasm of the cell, as these proteins are involved with the rearrangement of cytoskeleton and vacuolar sorting-associated protein. Svf1 protein is required for yeast cell survival under conditions of oxidative stress; thus, its higher abundance in 7B6-EV compared to 6B7-EV suggests diminished sensitivity to exogenous reactive oxygen species33. These data provide new information that helps to explain some of the different features (6B7 protective versus 7B6 non-protective) observed in vivo by the administration of these mAb to H. capsulatum infected mice11.
Pre-incubation of BMDM with EV prior to H. capsulatum challenge reduced the effector cells’ capacity to phagocytose yeast cells and, curiously, EV from H. capsulatum treated with the lowest concentration of both mAb were even more potent in inhibiting phagocytosis than control EV. The concentration-independency of mAb-mediated effects on EV was also seen in the proteomic data, where the abundance of some proteins were affected by the lowest concentration of the mAb but not by the higher. The treatment of BMDM with EV for 5 or 24 hours prior to the in vitro challenge with the yeast did not change the fungal uptake (data not shown). Therefore, we hypothesize that the Hsp60 present on the surface of the EV (from treated or untreated H. capsulatum) competes with Hsp60 present at the surface of the yeast for the CR3 site on the surface of BMDM, thereby inhibiting yeast cell recognition. However, after longer incubations, fungal EV are endocytosed by the macrophages16,34, increasing the availability of free CR3 on BMDM’s surface for the recognition and uptake of the yeast cells.
BMDM were less able to kill internalized H. capsulatum after the treatment with control EV, as well as with 6B7.20-EV. However, macrophages treated with 7B6.6-EV were even less able to kill H. capsulatum, supporting the concept that 7B6 is a non-protective or even a deleterious mAb. The intracellular killing inhibition on BMDM treated with 7B6.6-EV correlates with a marked reduction in the ROS generation in these BMDM, but also suggests that mechanisms other than ROS production may regulate the intracellular fate of the yeast.
Altogether, our data show that H. capsulatum is dynamically able to sense opsonization, and, in response to mAb, the yeast’s EV cargo is quantitatively and qualitatively changed in an isotype and concentration dependent manner. Moreover, these changes appear to affect the biology and pathogenicity of the fungus.
Methods
Histoplasma capsulatum
H. capsulatum G217B was purchased from the ATCC (ATCC, Cat# 26032) and the GFP strain (G217B background) was kindly provided by George S. Deepe (University of Cincinnati College of Medicine, Cincinnati, OH). Fungal cells were cultivated in Ham’s F-12 (Gibco, Cat# 21700-075) media supplemented with glucose (18.2 g/L), glutamic acid (1 g/L), HEPES (6 g/L) and L-cysteine (8.4 mg/L) at 37°C with constant shaking at 150 rpm. Fungal cultures were assessed for viability using Janus green 0.02%, and >99% of the yeast cells used were alive.
Cell Lines
All animal experiments were performed according to the Institute of Laboratory Animal Resources of the National Research Council guidelines and the Institutional Animal Care and Use Committee at the Albert Einstein College of Medicine approved the experimental methods. Balb/c mice (NCI, females aging 8–10 weeks) were euthanized and their femurs and tibias were excised and flushed with RPMI (Corning #10-040-CV) media to obtain cells. To obtain bone marrow-derived macrophages (BMDM), the cells were incubated with RPMI supplemented with 10% FBS and 20% of L929 (CLS Cat# 400260/p757_L-929, RRID:CVCL_0462) supernatant for 1 week, with media replacement at day 435. Alternatively, the human monocyte cell line THP-1 (CLS Cat# 300356/p804_THP-1, RRID:CVCL_0006) was treated with PMA (Sigma #P1585) 3 nM for 48 hours at 37°C to differentiate the cells into macrophages. THP-1 - differentiated macrophages were used in some experiments specified in the text.
mAb production
Under approval by the Einstein Institutional Animal Care and Use Committee, antibodies were generated in ascites. Briefly, mAbs were produced by the injection of 107 hybridoma cells into Pristane (Sigma-Aldrich, Cat# P2870) primed peritoneal cavities of ex-breeder Balb/c female mice (NCI). Alternatively, mAb were raised from hybridoma cells (6B7, 7B6)11 that were cultivated in DMEM (Corning, Cat# 10-017-CM) supplemented with 10% FBS, 10% NCTC-109 (Gibco #21340-039), 1% nonessential amino acids (Corning, Cat# 25-025-CI) and penicillin/streptomycin (Corning, Cat# 30-001-CI). Supernatants were harvested, filtered and concentrated. The concentrations of 6B7 and 7B6 mAb in ascites and supernatants were determined by ELISA using known concentrations of IgG1 or IgG2b standards, respectively12.
H. capsulatum treatment with protective (6B7) and non-protective (7B6) mAbs followed by extracellular vesicles purification
Exposure of H. capsulatum to mAb was performed according to a previously described protocol with minor modifications12,15,36,37. Briefly, H. capsulatum yeast cells were incubated with either 6 or 20 μg/mL of mAb 6B7 or 7B6. After 7 days of growth, the yeast cells were removed by centrifugation at 3,000 rpm for 10 minutes at 4 °C followed by filtration using a 0.45 μm pore filter38. EV purification from culture supernatant was achieved according to the protocol described by Rodrigues et al.39. Cell-free supernatant was concentrated in an Amicon ultrafiltration system with a 100-kDa cutoff membrane (Millipore #PBHK06210). The concentrated supernatant was further ultracentrifuged (Beckman Coulter Optima TLX) at 150.000 × g (52.000 rpm) using a TLA 100.3 rotor, (Beckman Coulter) for 1 hour at 4 °C. The EV were suspended in filtered PBS for proteomic analysis, and suspended in PBS with protease inhibitor cocktail (Roche) for dynamic light scattering analysis.
Protein and sterol quantification
The protein concentrations in EV samples were determined using BCA reagent (Thermo-Fisher #23227) measured using a nanodrop (ND-1000 Spectrophotometer, Thermo Scientific, USA). Sterol quantifications were performed using an Amplex Red kit (Molecular Probes #A12216) according to the manufacturer’s instructions (Life technologies, CA, USA).
Evaluation of enzymatic activities
EV suspensions with protein concentrations of 10 μg/mL were used to detect urease and laccase17. After addition of each enzyme reaction solution, the plates were stored at 37 °C and protected from light for 16 hours and then read using a spectrophotometer (BioTek, VT, USA). An enzyme reaction containing 1% peptone, 0.1% dextrose, 0.5% NaCl, 0.2% KH2PO4, 2% urea, and 0.0012% phenol red was used to evaluate urease activity, with plates read at 540 nm. To evaluate laccase, a solution with 12.5 mM of L-DOPA in PBS was utilized and the plates were read at 450 nm.
Proteomic analysis
The proteomic analysis was carried out as previously described in details12. Briefly, EV samples from two independent isolations (biological duplicates) of H. capsulatum treated or not with 20 μg/mL of either 6B7 or 7B6, were digested with trypsin and analyzed in an Ekspert nanoLC 400 system (Eksigent) connected to a 5600 TripleTOF mass spectrometer (AB Sciex). Peptide identification was performed with the Paragon tool of Protein Pilot software (AB Sciex) by searching against the H. capsulatum complete proteome set from Uniprot Knowledge Base. Peptide peak areas were extracted with Skyline40 and differential abundance analysis was done using the mapDIA software (http://mapdia.sourceforge.net)41. The results were compared to our previously published data of EV derived from cells treated with 6 µg/mL antibodies12. Protein functions were automatically annotated using the BlastKOALA tool42. Heatmaps were generated using Multiexperiment Viewer (MeV)43 and proteins with similar abundance profiles were clustered using the K-means and hierarchical methods.
Yeast Phagocytosis Assay
BMDM or THP-1-derived macrophages were plated in 12 well plates (5 × 105 cells/well) and allowed to adhere for one hour. After this incubation, cells were washed with PBS to remove non-adherent cells and incubated overnight at 370C at 5% CO2 for acclimatization. Macrophages were then incubated with 20 µg/mL (protein concentration) of EV 1 hour prior to the challenge with H. capsulatum-GFP (m.o.i. 1:5). After 1 hour of co-incubation, extracellular yeasts were removed by washing and then the macrophages were detached from the plate. Macrophages were analyzed by flow cytometry (FacScalibur – B&D) for fluorescence intensity and the percentage of infected cells determined. Data were normalized to the untreated (control) macrophages.
Yeast Killing Assay
To evaluate the intracellular killing, BMDM or THP-1-derived macrophages were treated with EV (20 μg/mL of protein concentration) for 1 hour prior to the challenge with H. capsulatum-GFP. After a 1 hour co-culture, the extracellular yeasts were removed by washing and the plates were incubated for additional 2 hours at 37°C and 5% CO2. The phagocytes were then lysed with distilled water and aliquots were plated onto BHI-agar plates. After 10 days the number of yeast colonies were counted and expressed as a percentage relative to the untreated group.
ROS production
To address ROS production, BMDM were plated (105 cells/well) in 96-well black plate. After the removal of non-adherent cells by washing, cells were loaded with H2DCFDA (10 μM). BMDM were then treated or not with EV (20 μg/mL of protein) for 30 minutes until the addition of H. capsulatum yeast cells. After a period of 2 hours, the plates were read in a SpectraMax M series.
Statistical analyses
Statistical analyses were performed by T-student or One-way ANOVA followed by Tukey test, using GraphPad Prism software.
References
Woods, J. P. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal genetics and biology: FG & B 35, 81–97, https://doi.org/10.1006/fgbi.2001.1311 (2002).
Nosanchuk, J. D. & Gacser, A. Histoplasma capsulatum at the host-pathogen interface. Microbes and infection 10, 973–977, https://doi.org/10.1016/j.micinf.2008.07.011 (2008).
Kauffman, C. A. Histoplasmosis: a clinical and laboratory update. Clinical microbiology reviews 20, 115–132, https://doi.org/10.1128/CMR.00027-06 (2007).
Allendoerfer, R. & Deepe, G. S. Jr. Infection with Histoplasma capsulatum: Host-fungus interface. Revista iberoamericana de micologia 15, 256–260 (1998).
Mihu, M. R. & Nosanchuk, J. D. Histoplasma virulence and host responses. International journal of microbiology 2012, 268123, https://doi.org/10.1155/2012/268123 (2012).
Bullock, W. E. & Wright, S. D. Role of the adherence-promoting receptors, CR3, LFA-1, andp150,95, in binding of Histoplasma capsulatum by human macrophages. The Journal of experimental medicine 165, 195–210 (1987).
Long, K. H., Gomez, F. J., Morris, R. E. & Newman, S. L. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. Journal of immunology 170, 487–494 (2003).
Strasser, J. E. et al. Regulation of the macrophage vacuolar ATPase and phagosome-lysosome fusion by Histoplasma capsulatum. Journal of immunology 162, 6148–6154 (1999).
Hwang, L. H., Seth, E., Gilmore, S. A. & Sil, A. SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryotic cell 11, 16–25, https://doi.org/10.1128/EC.05274-11 (2012).
Hilty, J., George Smulian, A. & Newman, S. L. Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Medical mycology 49, 633–642, https://doi.org/10.3109/13693786.2011.558930 (2011).
Guimaraes, A. J., Frases, S., Gomez, F. J., Zancope-Oliveira, R. M. & Nosanchuk, J. D. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infection and immunity 77, 1357–1367, https://doi.org/10.1128/IAI.01443-08 (2009).
Matos Baltazar, L. et al. Antibody Binding Alters the Characteristics and Contents of Extracellular Vesicles Released by Histoplasma capsulatum. mSphere 1, https://doi.org/10.1128/mSphere.00085-15 (2016).
Brown, L., Wolf, J. M., Prados-Rosales, R. & Casadevall, A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nature reviews. Microbiology 13, 620–630, https://doi.org/10.1038/nrmicro3480 (2015).
Nimrichter, L. et al. Extracellular Vesicle-Associated Transitory Cell Wall Components and Their Impact on the Interaction of Fungi with HostCells. Frontiers in microbiology 7, 1034, https://doi.org/10.3389/fmicb.2016.01034 (2016).
Albuquerque, P. C. et al. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cellular microbiology 10, 1695–1710, https://doi.org/10.1111/j.1462-5822.2008.01160.x (2008).
Vargas, G. et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cellular microbiology 17, 389–407, https://doi.org/10.1111/cmi.12374 (2015).
Rodrigues, M. L. et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic cell 7, 58–67, https://doi.org/10.1128/EC.00370-07 (2008).
Lee, J. et al. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrobial agents and chemotherapy 57, 2589–2595, https://doi.org/10.1128/AAC.00522-12 (2013).
Rivera, J. et al. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proceedings of the National Academy of Sciences of the United States of America 107, 19002–19007, https://doi.org/10.1073/pnas.1008843107 (2010).
Prados-Rosales, R. et al. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5, e01921–01914, https://doi.org/10.1128/mBio.01921-14 (2014).
Lee, W. H. et al. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Experimental & molecular medicine 47, e183, https://doi.org/10.1038/emm.2015.59 (2015).
Choi, S. J. et al. Active Immunization with Extracellular Vesicles Derived from Staphylococcus aureus Effectively Protects against Staphylococcal Lung Infections, Mainly via Th1 Cell-Mediated Immunity. PloS one 10, e0136021, https://doi.org/10.1371/journal.pone.0136021 (2015).
Nosanchuk, J. D. & Casadevall, A. The contribution of melanin to microbial pathogenesis. Cellular microbiology 5, 203–223 (2003).
Nosanchuk, J. D. et al. Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infection and immunity 70, 5124–5131 (2002).
Lee, I. R., Morrow, C. A. & Fraser, J. A. Nitrogen regulation of virulence in clinically prevalent fungal pathogens. FEMS microbiology letters 345, 77–84, https://doi.org/10.1111/1574-6968.12181 (2013).
Feder, V. et al. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. The FEBS journal 282, 1406–1418, https://doi.org/10.1111/febs.13229 (2015).
Stuehler, C. et al. Cross-protective TH1 immunity against Aspergillus fumigatus and Candida albicans. Blood 117, 5881–5891, https://doi.org/10.1182/blood-2010-12-325084 (2011).
Flannagan, R. S., Jaumouille, V. & Grinstein, S. The cell biology of phagocytosis. Annual review of pathology 7, 61–98, https://doi.org/10.1146/annurev-pathol-011811-132445 (2012).
Garfoot, A. L. & Rappleye, C. A. Histoplasma capsulatum surmounts obstacles to intracellular pathogenesis. The FEBS journal 283, 619–633, https://doi.org/10.1111/febs.13389 (2016).
Steenbergen, J. N., Nosanchuk, J. D., Malliaris, S. D. & Casadevall, A. Interaction of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infection and immunity 72, 3478–3488, https://doi.org/10.1128/IAI.72.6.3478-3488.2004 (2004).
Nosanchuk, J. D., Zancope-Oliveira, R. M., Hamilton, A. J. & Guimaraes, A. J. Antibody therapy for histoplasmosis. Frontiers in microbiology 3, 21, https://doi.org/10.3389/fmicb.2012.00021 (2012).
Silva, R. C. et al. Extracellular enolase of Candida albicans is involved in colonization of mammalian intestinal epithelium. Frontiers in cellular and infection microbiology 4, 66, https://doi.org/10.3389/fcimb.2014.00066 (2014).
Brace, J. L., Vanderweele, D. J. & Rudin, C. M. Svf1 inhibits reactive oxygen species generation and promotes survival under conditions of oxidative stress in Saccharomyces cerevisiae. Yeast 22, 641–652, https://doi.org/10.1002/yea.1235 (2005).
Oliveira, D. L. et al. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infection and immunity 78, 1601–1609, https://doi.org/10.1128/IAI.01171-09 (2010).
Filardy, A. A. et al. Proinflammatory clearance of apoptotic neutrophils induces an IL-12(low)IL-10(high) regulatory phenotype in macrophages. Journal of immunology 185, 2044–2050, https://doi.org/10.4049/jimmunol.1000017 (2010).
Reca, M. E. & Campbell, C. C. Growth curves with yeast phase of Histoplasma capsulatum. Medical mycology 5, 267–273, https://doi.org/10.1080/00362176785190511 (1967).
Goughenour, K. D., Balada-Llasat, J. M. & Rappleye, C. A. Quantitative Microplate-Based Growth Assay for Determination of Antifungal Susceptibility of Histoplasma capsulatum Yeasts. Journal of clinical microbiology 53, 3286–3295, https://doi.org/10.1128/JCM.00795-15 (2015).
Wolf, J. M. & Casadevall, A. Challenges posed by extracellular vesicles from eukaryotic microbes. Current opinion in microbiology 22, 73–78, https://doi.org/10.1016/j.mib.2014.09.012 (2014).
Rodrigues, M. L. et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryotic cell 6, 48–59, https://doi.org/10.1128/EC.00318-06 (2007).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968, https://doi.org/10.1093/bioinformatics/btq054 (2010).
Gil-Bona, A. et al. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. Journal of proteome research 14, 142–153, https://doi.org/10.1021/pr5007944 (2015).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol 428, 726–731, https://doi.org/10.1016/j.jmb.2015.11.006 (2016).
Chu, V. T., Gottardo, R., Raftery, A. E., Bumgarner, R. E. & Yeung, K. Y. MeV + R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol 9, R118, https://doi.org/10.1186/gb-2008-9-7-r118 (2008).
Acknowledgements
Parts of this work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA; and the Bindley Bioscience Center, Discovery Park, Purdue University, West LaFayette, IN. L.M.B. was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) process no. 3226-13-1. J.D.N. is partially supported by NIH 3R37AI033142, and J.D.N. and E.S.N. are partially supported by NIH R21 AI124797. L.N. is supported by grants from CNPq (National Counsel of Technological and Scientific Development, Brazil) and FAPERJ (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Brazil).
Author information
Authors and Affiliations
Contributions
L.M.B., D.Z.-M., L.N., E.S.N. and J.D.N. designed research; J.D.N. supervised the study; L.M.B., D.Z.-M. and E.S.N. performed research; L.M.B., D.Z.-M., M.C.B., L.N., H.C., E.S.N. and J.D.N. analyzed data; H.C., E.S.N. and J.D.N. contributed with new reagents/analytic tools; L.M.B., D.Z.-M., E.S.N. and J.D.N. wrote the paper; E.S.N. and J.D.N. provided lab space, instrumentation and funding; All authors edited and gave critical input on the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baltazar, L., Zamith-Miranda, D., Burnet, M.C. et al. Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Sci Rep 8, 8065 (2018). https://doi.org/10.1038/s41598-018-25665-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25665-5
This article is cited by
-
Yeast strains isolated from fermented beverage produce extracellular vesicles with anti-inflammatory effects
Scientific Reports (2024)
-
Characteristics and potential clinical applications of the extracellular vesicles of human pathogenic Fungi
BMC Microbiology (2023)
-
Extracellular Vesicles From Sporothrix brasiliensis Yeast Cells Increases Fungicidal Activity in Macrophages
Mycopathologia (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.