Abstract
Unilateral eyestalk ablation in the female black tiger shrimp Penaeus monodon is commonly employed to induce ovarian maturation. However, the importance of complementing this practice with the provision of live feed supplement (such as polychaetes) has not been emphasized in previous studies. Indeed, it has been less emphasized that female broodstock must be fed with live feeds such as polychaetes for this practice to be effective. While the effects of eyestalk ablation have been thoroughly studied in various aspects, the synergistic effects of feeding with live feeds and the ablation have never been elucidated at a transcriptome-wide level. With recent advances in the next-generation sequencing platforms, it is now possible to investigate the effects of eyestalk ablation and live feeds at the transcriptomic levels. This study employed both short-read Illumina RNA sequencing and long-read Pacific Biosciences (PacBio) isoform sequencing (Iso-seq) to generate the first high-quality ovarian reference transcriptome in P. monodon. This novel assembly allowed us to dissect the effects of feeds and eyestalk ablation and reveal their synergistic effects at the transcriptomic level through the regulation of important genes involved in fatty acid regulation, energy production, and hormone-mediated oocyte maturation pathways. The synergistic effects between the polychaete feeding and the eyestalk ablation in the process of ovarian maturation in black tiger shrimp suggest that without having proper nutrients from the polychaetes, female broodstock might not be ready to develop its ovary. However, even with proper nutrients, the eyestalk ablation is still necessary to perhaps manipulate the female endocrine of the black tiger shrimp. These findings shed the light on molecular mechanisms and key molecular pathways that lead to successful ovarian maturation.
Similar content being viewed by others
Introduction
The black tiger shrimp (Penaeus monodon) is the second most widely cultured shrimp species in the world (FAO 2016). Albeit high demand worldwide, the shrimp production level remains stagnant mainly due to the outbreak of diseases and lack of selective breeding programs. The future success of the shrimp industry will depend upon increasing supplies of healthy, high-quality seed for stocking ponds and decreasing reliance on wild broodstock, which might potentially be a disease carrier and have seasonal variation1. To overcome these challenges, it is vital to be able to enhance reproductive maturation of captive broodstock.
Ovarian maturation is a well-orchestrated and complex molecular process that requires the regulation of large sets of genes to ensure proper oocyte development for embryogenesis. In penaeid shrimp, the process can be categorized into four stages: (I) previtellogenesis, (II) vitellogenesis, (III) early cortical rod, and (IV) late cortical rod2. Numerous factors and methods have been investigated to improve reproductive maturation in female black tiger shrimp. Dietary choices have been shown to be important for the maturation. Although nutritional requirements for shrimp reproduction are poorly defined, live feeds such as polychaetes, squid, and mussels are generally used to prepare broodstock before mating because they are nutritionally rich in proteins, lipids, polyunsaturated fatty acids (PUFA)3,4, and sex steroid hormones, such as progesterone and its derivatives5. Dietary PUFAs have been reported to benefit ovarian development in several crustacean species, such as the Pacific white shrimp Litopenaeus vannamei6, the Chinese mitten crab Eriocheir sinensis7, the mud crab Scylla serrata8 and P. monodon9. However, in P. monodon, the maturation diet alone could not induce female maturation to the desirable degree, and unilateral eyestalk ablation is still necessary10.
Eyestalk ablation is generally practiced to induce ovarian maturation in shrimp farming11,12. This technique has been reported to enhance the ovarian maturation by eliminating gonad-inhibiting hormone (GIH)/vitellogenesis-inhibiting hormone (VIH) secreted from the X-organ sinus gland complex within eyestalks13. From this knowledge, GIH-specific double-stranded RNA was used experimentally to silence GIH expression, which in turn increased the expression level of vitellogenin gene, ovarian maturation and spawning in both domesticated and wild female broodstock P. monodon shrimp14. However, the brooders used in the study were also fed with polychaetes and squids. Besides the GIH removal hypothesis, eyestalk removal has been linked to reduced light intensity and thereby inducing ovarian maturation in the banana prawn P. merguiensis15. The exact mechanism by which the eyestalk ablation induces ovarian maturation remains inconclusive.
Previously, we employed cDNA microarray to investigate the molecular and physiological effects of unilateral eyestalk ablation leading to ovarian maturation in polychaete-fed female P. monodon and discovered the up-regulation of genes in several crucial pathways, namely the gonadotropin-releasing hormone (GnRH) signaling pathway, calcium signaling pathway, and progesterone-mediated oocyte maturation pathway16. However, it was not clear whether the reported changes in transcript accumulation of genes in those pathways were affected by the eyestalk ablation, the polychaete diet, or both. Moreover, with the limitation of cDNA microarray technique, transcripts excluded from the probes on the microarray were not examined.
In this study, we employed the next-generation sequencing to dissect the effects of eyestalk ablation and feeds on gene expression in domesticated P. monodon female broodstock by comparing changes in transcript accumulation between two groups of broodstock fed with either pellet or polychaetes before and after eyestalk ablation.
Results
Effects of feeds and eyestalk ablation on ovarian maturation
To examine the effects of feeding and eyestalk ablation on ovarian maturation of domesticated female black tiger shrimp broodstock, shrimps were fed with either broodstock pellets or live polychaetes for four weeks before an eyestalk ablation was performed (Fig. 1). Both feeds could not induce ovarian maturation over the 4-week feeding period as evidenced by the pale white color of ovaries (Fig. 1A) and the insignificant changes in gonadosomatic index (GSI), a known indicator of ovarian maturation in the female black tiger shrimp (0.57 ± 0.18% to 0.54 ± 0.18% for the pellet-fed group and 0.57 ± 0.18% to 0.72 ± 0.21% for the polychaete-fed group; Fig. 1B, Supplementary Table S1).
Ovarian maturation after unilateral eyestalk ablation in the shrimp fed with pellet or polychaetes. (A) Representatives of ovaries from different time points. (B) Gonadosomatic Index (GSI) of the shrimp at different time points. Error bars represent standard derivation. Different letters indicate significant differences among time points of the same feed group (p < 0.05, ANOVA Duncan test). Asterisks indicate significant differences between the two feed groups at the same time point (p < 0.05, t-test).
After the eyestalk ablation, ovaries of the two groups matured differently. The ovaries of the pellet-fed group did not develop further than previtellogenic stage (stage I, see Materials and Methods for definition of ovarian stages) as they remained white-yellow in color and small in sizes with the GSIs of 0.61 ± 0.20%, 0.48 ± 0.08%, and 0.78 ± 0.31% for Day 1, 4, and 9 post ablation, respectively. On the other hand, the polychaete-fed shrimp were able to reach the reproductively mature stage after the eyestalk ablation as their ovaries continually became larger and eventually turned greenish by Day 9 post ablation. Their GSIs started from 0.72 ± 0.21% and significantly increased to 1.26 ± 0.57%, 3.61 ± 1.14%, and 5.88 ± 0.58% after eyestalk ablation at Day 1, 4, and 9, respectively (Fig. 1B, Supplementary Table S1).
P. monodon ovarian transcriptome assembly and annotation
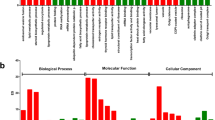
To generate the reference ovarian transcriptome sequences for black tiger shrimp, we applied both short-read Illumina sequencing and long-read Pacific Biosciences (PacBio) isoform sequencing (Iso-seq). The depth coverage of Illumina sequencing allowed us to capture the sequences of rare transcripts (Supplementary Table S2) while the long PacBio reads enabled the identification of full-length transcripts (Supplementary Table S3). After processing raw read data from the PacBio RS II, we obtained a total of 546,473 reads of insert (ROIs) totaling 18.82 Gb from 8 SMRT cells (Table 1; Supplementary Table S3). Of the 546,473 ROIs, 26,609 were high-quality, full-length non-chimeric transcripts identified based on the presence of 5′ and 3′ cDNA primers and a polyA tail signal preceding the 3′-primer. The sizes of the ROIs ranged from 10 to 15,000 with an average read length of 2,087 and an N50 of 2,454 nucleotides. Short-read RNA sequencing was also carried out on the Illumina NovaSeqTM 6000 and yielded 1,235,809,734 raw reads, totaling 187.92 Gb. After a de novo transcriptome assembly using Trinity v2.2.017, we obtained 398,088 assembled transcripts ranging from 201 to 13,211 nucleotides. Of which, 204,819 sequences had a length of 300 nucleotides or longer (Table 1). To enhance the coverage of the reference transcriptome assembly, we combined high-quality, full-length transcripts from PacBio Iso-seq experiment with Trinity-assembled transcripts longer than 300 nucleotides. After clustering these sequences at 95% identity, we obtained a final set of 33,277 non-re dundant transcripts with an average contig length of 917 nucleotides and an N50 length of 1,728 nucleotides (Table 1). We investigated the coverage of the benchmarking universal single-copy orthologs (BUSCO)18 to assess the completeness of the gene space in the assembly. Even though we only examined transcripts deriving from ovary tissues of the black tiger shrimp, 92.7% of the assembly was identified as ‘complete’ (i.e., the length of the recovered genes are within two standard deviations of the BUSCO group mean length18), suggesting that the assembly is relatively complete with respect to the gene space. Gene ontology (GO) annotations describing biological processes, molecular functions, and cellular components were retrieved for 15,480 sequences (46.52% of the assembly; Supplementary Fig. S1). The most prevalent GO term associated with biological processes were oxidation-reduction process (239) followed by protein phosphorylation (157), translation (147), proteolysis (146), regulation of transcription (139), membrane transport (111) and signal transduction (103; Supplementary Fig. S1, Table S4). Among genes annotated to various molecular functions, the largest category was ATP binding (597), followed by metal ion binding (286) and nucleic acid binding (271), another ten subcategories with 100 or more terms (Supplementary Fig. S1, Table S4).
Differential gene expression profiles
Feed effects
After the 4-week feeding trial, only a small portion of total transcripts was differentially expressed in both feed groups, and each type of diet regulated different sets of genes (Fig. 2A). For the pellet-fed group, 20 transcripts were differentially expressed, whereas 12 transcripts were for the polychaete-fed group. The pellet-fed group significantly induced mitochondrial assembly of ribosomal protein, furin-like protease, replication protein and transcription factor, all of which appeared to be involved in regular cell maintenance. On the other hand, the polychaete-fed group differentially induced genes related to oogenesis such as vitellogenin, collagen alpha-1V chain-like, and heme-binding protein 2-like, suggesting that the polychaete-fed shrimp were able to prepare for ovarian maturation process after obtaining nutrients necessary for reproduction provided by polychaetes.
Eyestalk ablation effects
After the eyestalk ablation, the two feed groups exhibited almost entirely different expression profiles, and the two diets regulated different sets of genes (Supplementary Fig. S2). The number of differentially expressed genes (DEGs) was higher for the polychaete-fed group than the pellet-fed group throughout the ablation time course (Fig. 3A). While the pellet-fed group differentially expressed most number of genes (77 transcripts) at Day 1 after the ablation, the polychaete-fed group differentially expressed most number of genes (1,522 transcripts) at Day 4 (Fig. 3), coinciding with the time when the most noticeable change in ovarian physiology occurred (Fig. 1A).
To examine transcriptomic responses after eyestalk ablation, DEGs at each time point were identified. From 11,694 non-redundant annotated transcripts, 1,559 features that were significantly up- or downregulated (p-adjusted <0.05) in at least one time point were clustered according to their expression patterns (Fig. 4B). We subsequently clustered them based on their expression patterns and selected groups of genes that were affected by feed and eyestalk ablation. A total of 442 features exhibited changes in their expression level >1.5 folds (Supplementary Table S5), and selected features with known functions were plotted in groups according to their similar expression patterns (Fig. 4C).
Gene expression analysis by RNA sequencing. Transcript levels in ovaries after a 4-week feeding trial (T1) were compared to before the trial (T0). Transcript levels in ovaries after eyestalk ablation at Day 1, 4 and 9 (T2, T3, and T4, respectively) were compared to those from non-ablated broodstock (T1). (A) A total of non-redundant and annotated 11,694 features were obtained from RNA sequencing. (B) Hierarchical clustering analysis of the 1,559 features with significant differences (p-adjusted < 0.05) in at least 1 time point of 8 time points. Transcript levels in ovaries after eyestalk ablation at Day 1, 4 and 9 (T2, T3, and T4, respectively) were compared to those from non-ablated broodstock (T1). (C) Genes that exhibited changes in their expression level > 1.5 folds (442 features) were clustered into Cluster I–II exhibiting various differentially expressed patterns as effects of diets and eyestalk ablation: Upregulation in pellet-fed group (I), Upregulation in polychaete-fed group (II) with cluster IIa contains genes with upregulation before (T1) and after (T2-T4) the ablation, cluster IIb contains genes with upregulation after the ablation (T2, Day 1) until T4 (Day 9), cluster IIc contains genes with upregulation at T3 (Day 4) after the ablation, and cluster IId contains genes with upregulation at T4 (Day 9) after the ablation. Transcripts in blue letters were further characterized by quantitative real-time PCR (Supplementary Fig. S4).
The eyestalk ablation affected almost totally different sets of genes between the two feed groups with negligible number of co-expressed genes in both time courses (Fig. 2B). For the pellet-fed group, the early response at Day 1 seemed to induce genes involved in stress recovery such as cytoskeletal and cell repair (actin, catenin alpha, acidic mammalian chitinase), homeostasis maintenance (mitochondrial carrier Rim2), and protein repair (adenosine kinase2, tyrosine-tRNA ligase, mitochondrial; Fig. 4). At Day 4 after the ablation, the pellet-fed group differentially induced farnesoic acid O-methyltransferase (FAMeT), Niemann-Pick C1 protein-like, and rho GTPase-activating protein 6 isoform X3, while the well-known ovarian maturation marker gene, vitellogenin (Fig. 4, Cluster IIa), was only differentially induced at Day 9 after the eyestalk ablation.
For the polychaete-fed group, a large number of transcripts were differentially expressed after the eyestalk ablation, which can be categorized according to their expression patterns (Fig. 4). The first pattern includes transcripts that were upregulated after four weeks of polychaete feeding and their levels of expression continually increased after the eyestalk ablation (Fig. 4, Cluster IIa). These genes are vitellogenin, collagen alpha-1V chain-like, and heme-binding protein 2-like. The second group of genes were upregulated early (Day 1), and most of them remained upregulated until the end of the time-course experiment after the ablation (Fig. 4, Cluster IIb). This group includes fatty acid regulatory genes (fatty acid synthase, very low density lipoprotein receptor-like isoform X1, stearoyl-CoA desaturase 5-like, cation-dependent mannose-6-phosphate receptor-like, mitochondrial pyruvate carrier1-like), mitosis and meiosis regulation (mitotic checkpoint protein BUB3-like isoform X3, meiosis regulator and mRNA stability factor 1-like), purine regulatory genes (inosine-5′-monophosphate dehydrogenase 1b-like isoform X1 and 3-phosphoinositide-dependent protein kinase 1-like), protein degradation/modification via ubiquitination (E3 ubiquitin-protein ligase RING1-like, ubiquitin-like protein ATG12, histone H1.3-like, histone deacetylase complex subunit SAP30 homolog, energy production genes (NADH dehydrogenase subunit 1-4, cytochrome b mitochondrion, cytochrome c oxidase subunit I, II, III mitochondrion, ATP synthase), and oogenesis genes (polehole-like protein, vasa-like protein and vitelline membrane outer layer protein 1 homolog; Fig. 4: Cluster IIb).
At Day 4 after the ablation, several genes belonging to a hormone-mediated ovarian maturation and calcium signaling pathways such as progestin membrane receptor component 1 (PGMRC1), estrogen sulfotransferase, crustacean hyperglycemic hormone 3 precursor, calmodulin, guanine nucleotide-binding protein-like 1 (Gq/11) and 45 kDa calcium binding protein-like were differentially induced. In addition, transport proteins (transport and Golgi organization protein 6-like isoform X3), fatty acid regulatory genes (elongation of very long chain fatty acids protein 6), collagen gene and its receptor gene (collagen alpha-3(IX) chain-like, and integrin beta-PS-like isoform X1), and cellular development (mitogen-activated protein kinase kinase (MAPKK)) were induced at Day 4 (Fig. 4: Cluster IIc). For Day 4 to Day 9 after the ablation, vitellogenesis genes (vitellogenin receptor and NPC intracellular cholesterol transporter 2 homolog a-like), cellular development genes (innexin 2-like isoform X2, rab GTPase and CDK5 and ABL1 enzyme substrate 1-like), osmotic regulatory gene (aquaporin 1), reproduction-related gene (vigilin) and gluconeogenesis gene (phosphoenolpyruvate carboxykinase) were induced (Fig. 4: Cluster IIc). For the later time point at Day 9, cathepsin C, kinesin-like protein KIF18A, calcineurin-binding protein cabin-1-like, DNA damage-inducible transcript 4-like protein, multiple inositol polyphosphate phosphatase 1-like, transport protein Sec. 24 C and calcium-activated chloride channel-regulator 2-like were induced (Fig. 4: Cluster IId).
From these 442 differentially expressed genes, pathway analysis was performed to identify enriched pathways and the most enriched pathway was found to be very general “metabolic pathway” (Supplementary Fig. S3). However, the second most enriched pathway namely the gonadotropin-releasing hormone (GnRH)/MAPK signaling pathway presented relevant biological functions to the reproductive maturation. Therefore, expression levels of these transcripts in this pathway were further investigated and potentially found to be involved in the shrimp reproductive maturation (Fig. 5).
Differential expressed genes in the hormone-mediated ovarian maturation pathways in the polychaete-fed group. Light blue box, yellow box and white box represent receptor genes, genes were found in this study, and genes known to be in this pathway, respectively. The heatmap bars under each gene in the pathway reflect expression change from RNA-seq result after a 4-week feeding trial (T1) compared to before the trial (T0), after eyestalk ablation at Day 1, 4 and 9 (T2, T3, and T4, respectively) compared to those from non-ablated broodstock (T1). GnRH: Gonadotropin releasing hormone, GnRHR: GnRH receptor, G q/11: guanine nucleotide-binding protein G(q) subunit alpha, Plcβ: phospholipase C beta, IP3: inositol-1,4,5-triphosphate, IP3R: IP3 receptor, Calr: calreticulin, Calm: calmodulin, CABIN1: calcineurin-binding protein cabin-1-like, Cdc20: cell division cycle 20, APC/C: Anaphase-promoting complex, CamKII: calmodulin kinase II, Cdc25C: cell division cycle 25C, PGMRC1:progestin membrane receptor component 1, G Gi α: guanine nucleotide-binding protein G(i) subunit alpha, AC: adenylate cyclase, Plkkl: Polo-like kinase I, Cdc2: cell division cycle 2, CycB: cyclinB, MPF: maturation-promoting factor, MAPKK: mitogen-activated protein kinase kinase, PAK2: serine/threonine-protein kinase PAK 2-like, ELK1: ETS-like protein pointed isoform X5.
Validation of gene expression by quantitative real-time PCR (qPCR) analysis
To confirm the expression patterns of selected transcripts with differential expression levels in the RNA-seq result, real-time qPCR was performed for nine transcripts with significant expression levels in polychaete-fed group, especially after the eyestalk ablation (Supplementary Fig. S4). All examined transcripts were differentially expressed between pellet-fed and polychaete-fed groups after the 4-week feeding trial. As observed in the RNA-seq result, vitellogenin was expressed highly in polychaete-fed group after the 4-week trial. After the eyestalk ablation, all of these transcripts were expressed higher in the polychaete-fed group than the pellet-fed group, with cytochrome c oxidase and vitellogenin being the most obvious transcripts.
Discussion
Poor ovarian maturation in shrimp domestication hampers further growth and viability of the industry. While several methods such as boosting with live feeds and unilateral eyestalk ablation have been used to induce the maturation, the molecular effects of these methods have not been investigated in detail. This study employed transcriptomic analyses to elucidate the effects of live feed diet and eyestalk ablation on ovarian maturation and reveal important genes and pathways involved. The availability of high-quality ovarian transcriptome assembly is crucial for understanding the underlying transcriptional regulations triggered by the interactions between diet and eyestalk ablation. Previously, we employed 454 pyrosequencing technology to obtain transcriptomic information on reproduction of the P. monodon ovary and testis, and found several novel sets of transcripts19. However, the transcriptomic information obtained was of short-read sequences due to the limitation of the sequencing technology employed at that time. In this work, we reported the first high-quality reference ovarian transcriptome in P. monodon assembled from both Illumina short-read sequences and PacBio long-read sequences, the latter of which enabled us to capture full-length transcripts. Our assembly has a much higher proportion of annotated sequences (46.52%) compared to the number reported in a previous study (17.83%)20. Future expression studies and genome sequencing project in black tiger shrimp will greatly benefit from the annotated transcriptome assembly reported here.
Ovarian maturation in penaied shrimp requires coordination of large sets of genes for dramatic physiological changes into four stages: (I) previtellogenesis, (II) vitellogenesis, (III) early cortical rod, and (IV) late cortical rod2. This study shows that without eyestalk ablation, the P. monodon broodstock was unable to develop pass the previtellogenesis stage regardless of the feed given. However, eyestalk ablation was able to induce ovarian maturation only in the group fed with polychaetes, indicating that there was a synergy between polychaetes and ablation effects. The significant changes in GSI and physiology of the polychaete-fed ovaries after the ablation suggested that feeding with polychaetes helped prepare the broodstock before the eyestalk ablation.
Feed effects
Nutritional requirements for shrimp reproduction are poorly defined. Generally, live feeds such as polychaetes are used to prepare broodstock before mating, but their nutritional value can vary with species, season of collection and life stage21. Their use is also labor-intensive, expensive and affects water quality22. The replacement of live feeds with dry feeds for shrimp reproduction will not be possible without understanding the molecular effects of how these live feeds prepare broodstock during ovarian maturation.
From the transcriptomic data, diets showed strong influences on gene regulation. The two examined types of feed regulated totally different sets of genes throughout the experiment as evidenced by an almost complete absence of co-expressed DEGs in both feed groups, suggesting that the pellet and polychaete utilized different molecular pathways. For the pellet-fed group, genes with altered expression levels belong to regular cell maintenance, and there was no significant alteration of expression levels for genes related to reproduction. Oppositely, the polychaete-fed group differentially expressed a large number of important genes whose functions are relevant to the preparation for ovarian maturation.
Polychaetes are high in polyunsaturated fatty acids (PUFAs), which have been reported to enhance ovarian maturation in many crustacean species such as the Pacific white shrimp Litopenaeus vannamei, the Chinese mitten crab Eriocheir sinensis, the mud crab Scylla serrata and P. monodon7,8,9,23. This study reveals the benefits of polychaete at the molecular level, which are highlighted by a strong induction of an oogenesis gene marker, vitellogenin, just after four weeks of polychaete feeding and prior to the eyestalk ablation (Fig. 4: Cluster IIa). Vitellogenin, also known as apolipophorin, is a precursor for the synthesis of a major egg yolk protein vitellin through vitellogenesis process24. This protein provides nutrition required for crustacean embryo development25. Hepatopancreas and ovary are common sites for yolk biosynthesis in several penaeid shrimps such as Litopenaeus vannamei26, Penaeus semisulcatus27 and Penaeus monodon28. While the level of vitellogenin transcripts was significantly induced in the polychaete-fed group within the first 24 hours after the removal of the eyestalk, the same induction was not observed until Day 9 for the pellet-fed group (Fig. 4: Cluster IIa). Coordination of reproduction and yolk biosynthesis is achieved via the endocrine system, which was partly located in the eyestalk29,30. While it is not surprising that the vitellogenin transcript was induced upon removing GIH through eyestalk ablation, it is worth noting that its expression level was induced even before the ablation in the polychaete-fed group, providing the evidence to support that ovarian maturation not only depends on the release of hormone, but also on the readiness primed by specific nutrition obtained from the polychaetes. Moreover, the fact that the vitellogenin transcript was upregulated at Day 9 after the ablation in the pellet-fed group but did not result in ovarian maturation also suggests that the completion of the maturation process requires various factors to work together.
In addition to the induction of vitellogenin, polychaete feeding differentially induced heme-binding protein and collagen alpha-1 even before the eyestalk ablation. While both of these genes have not previously been studied in P. monodon, they have been linked to reproduction in other organisms. Heme-binding protein encodes a specific yolk protein in the insect Rhodnius prolixus, and its uptake in the ovary was modulated by eicosanoids31. Eicosanoids, hormone-like mediators of reproduction, have been reported to be involved in female reproduction of numerous crustaceans such as Daphnia magna32. Eicosanoids or prostanoids play crucial roles in regulation of female reproductive maturation33,34. Injection of some PUFA derivatives such as prostaglandin F(2alpha) and prostaglandin E(2) significantly increased an ovarian index and oocyte diameter in fresh water crabs35. PUFAs in polychaetes are the precursors of eicosanoids; thus, it is not surprising that heme-binding protein was induced after feeding polychaetes for four weeks. Similarly, collagen alpha-1 was induced before eyestalk ablation in the polychaete-fed group. Although the roles of collagen for ovarian maturation has not been studied in the penaied shrimp, collagens are a major component of the extracellular matrix, which regulates follicle maturation and a process of explosive follicle growth called folliculogenesis in several model organisms36. Various types of collagens were present throughout the surface epithelium, follicular compartments, stroma, and granulosa cells of mouse ovary37.
Synergistic effects from diets and eyestalk ablation
The most interesting DEGs were those induced in the polychaete-fed group after the eyestalk ablation but not in the pellet-fed group. We hypothesized that those transcripts were up- or downregulated as a result of synergistic effects from polychaete feeding and eyestalk ablation. While most of those genes were involved in fatty acid regulation, energy production, and hormone-mediated oocyte maturation pathways, there were several genes whose functions have previously been linked to various biological processes worth examining in detail.
Fatty acid regulation genes
Given polychaete’s richness in PUFAs, it is not surprising to find several genes involved in fatty acid regulatory networks differentially specifically induced in the polychaete-fed group, especially after the eyestalk ablation. Previously, genes in the fatty acid regulation pathways have been shown to be highly expressed in mature ovaries of P. monodon fed with polychaetes after eyestalk ablation38. Fatty acids and dietary lipids are important for vitellogenesis as yolk proteins are composed mainly of lipoglycoproteins39. The importance of fatty-acid rich feed to the ovarian development in crustaceans has previously been demonstrated. In L. vannamei, the higher fatty acid content in feed led to a higher vitellogenin concentration in hemolymph40, whereas dietary lipids influenced gene expression of fatty acid regulatory pathways including fatty acid beta-oxidation, metabolism, synthesis and transport in the Chinese mitten crab E. sinensis and the Pacific white shrimp L. vannamei41,42.
Energy production genes
The induction of energy-related genes such as ATP synthase, cytochrome, NADH dehydrogenase, phosphoenolpyruvate carboxykinase, glucose receptor seems to be a result of synergistic effect from the polychaete feeding and the eyestalk ablation (Fig. 4: Cluster IIb). The importance of energy to reproduction has been evidenced by the fact that a large amount of lipids stored in the hepatopancreas are used for energy generation and directly transferred to the ovary during ovarian development43.The lipid stored in eggs will be mobilized for energy production during the course of embryogenesis44.
Hormone-mediated oocyte maturation genes
Another interesting group of genes that were upregulated specifically in the polychaete-fed group after the eyestalk ablation includes genes encoding gonadotrophin releasing hormone (GnRH) signaling, calcium signaling and progesterone pathways such as calmodulin, 45 kDa calcium-binding protein-like, mitogen-activated protein kinase kinase (MAPKK), progestin membrane receptor component 1 (PGMRC1), and progesterone-induced-blocking factor (Fig. 4: Cluster IIc, Supplementary Table S5). Moreover, cathepsin C was found to be highly induced at the later time point after the eyestalk ablation in the polychaete-fed group, which coincides with the final stage of oocyte maturation indicated by the GSI and ovarian morphology. Cathepsin C, known as dipeptidyl peptidase I (DDPI) or dipeptidyl transferase, is a lysosomal cysteine protease that is involved in the final stages of oocyte maturation in kuruma prawn (Marsupenaeus japanicus)45. It plays physiological roles in intracellular protein degradation and has been postulated to facilitate cortical rod expulsion at the exposure of seawater. Our expression data agree well with the literature, suggesting that cathepsin C plays a vital role in the final stage of the P. monodon oocyte maturation. The induction of these genes confirms our finding from previous studies using cDNA microarray to decipher affected pathways after eyestalk ablation16 and 454 pyroseqeuncing to reveal pathways and genes related to reproduction of P. monodon19. The expression patterns of these gene were validated using real-time qPCR. Most of the results from the qPCR agreed with those from the RNA-seq.
Ubiquitin, protein degradation or modification genes
Genes involved in ubiquitin-mediated proteolysis, protein degradation or modification (importin subunit alpha-4, E3 ubiquitin-protein ligase RING1-like, ubiquitin-like protein ATG12, histone H1.3-like, histone deacetylase complex subunit SAP30 homolog) were up-regulated after eyestalk ablation in the polychaete-fed group. Protein ubiquitination plays a role in protein refolding and is involved in the control of a multitude of processes such as cell differentiation, the cell cycle, responses to stress, chromatin modification in the context of DNA repair, gene silencing, as well as, gametogenesis in various organisms46. For histone genes, it is not surprising to see their involvement during the ovarian maturation in this study. In Caenorhabditis elegans, a histone deacetylase, HDA-1 forms a protein complex with nuclear autoantigenic sperm protein (NASP) and a zinc finger transcription factor, TRA-4 to promote female development47, while NASP has been linked to female reproductive maturation in P. monodon48.
Other genes
First, crustacean hyperglycemic hormone (CHH) is a neuropeptide that is synthesized, stored and released by eyestalk structure in crustaceans49. From our result, crustacean hyperglycemic hormone 3 (CHH3) precursor was up-regulated after eyestalk ablation in the polychaete-fed group. When we performed the sequence alignment (BLASTX) of our CHH3 precursor to the Genbank nr database, the best hit returned was CHH3 precursor from L. vannamei. When we aligned our CHH3 precursor with previously published Pem-CHH1, Pem-CHH2 and Pem-CHH3 sequences of P. monodon50, the percent identity obtained was very low (<20%) and the aligned portions were also small. Based on these results, it is unlikely that our CHH3 precursor is the same as Pem-CHH3 (or Pem-CHH1 and Pem-CHH2). It is probably a new member of the CHH family that has not been reported.
It is a member of a structurally related peptide family, which includes molt-inhibiting hormone (MIH), gonad-inhibiting hormone (GIH) and mandibular organ-inhibiting hormone (MO-IH)50. Reported to be involved in several physiological processes such as edysteroid production, lipid metabolism, ovarian physiology and osmoregulation, CHH’s main function is to regulate sugar levels in the hemolymph and glycogen metabolism. Given that the upregulation of this gene was observed after the eyestalk ablation only in the polychaete-fed group (Fig. 4: Cluster IIc), we hypothesize that this gene helps boost the sugar level in ovary tissues to provide the energy necessary for the ovarian maturation.
Polehole exhibiting similar expression patterns within the synergistic gene group may play important roles for ovarian maturation. In Drosophila, Polehole located on the oocyte surface mediates the assembly of the eggshell participating in the Torso RTK signaling pathway that specifies the terminal regions of the embryo and crosslinks vitelline membrane product51. In P. monodon, a transcript that significantly matched the polehole precursor was characterized and found to be expressed specially in ovary, highly expressed after eyestalk-ablation and localized in the ooplasm of oocyetes, suggesting its important function in oocyte development and maturation52. However, its detailed molecular mechanism needs to be further explored.
Conclusion
Our RNA-seq results strongly suggest that there are synergistic effects between the polychaete feeding and the eyestalk ablation in the process of ovarian maturation in black tiger shrimp. Without having proper nutrients from the polychaetes, female broodstock might not be ready to develop its ovary; however, even with the proper nutrients, the eyestalk ablation is still necessary to perhaps manipulate the female endocrine of the black tiger shrimp. The synergy seems to be the induction of important groups of genes, namely energy production genes, endocrinological genes, fatty acid regulatory genes, and oogenesis genes. These findings shed to light on molecular mechanisms and key molecular pathways that lead to successful ovarian maturation.
Material and Methods
Experimental design, sample collection, and RNA extraction
The shrimp culture conditions were followed a previously published work16. Female domesticated P. monodon broodstock (9-month-old) and live sand polychaetes Perinereis nuntia (6-month-old) were cultured at the Shrimp Genetic Improvement Center (SGIC, Surat Thani, Thailand). Brooders were maintained at a biosecure station containing seawater pumped from the Gulf of Thailand at a salinity of 30 ppt with water temperature at around 27 °C, and acclimated in tanks with 2 m in diameter for 2 weeks. The brooders were separated into two groups (n = 46 each): pellet-fed (Pe) and polychaete-fed (Po) groups. Pellet (Pe) and polychaete (Po) groups were fed five times/day (8:00, 11:00, 14:00, 16:30, and 22:00 h). The amount of feed/shrimp weight/day was 2% broodstock pellets (Charoen Pokphand Foods PCL) for Pe group and 7% polychaetes for Po group.
Ovaries of the brooders were collected at the beginning of the feeding trial (T0), 4 weeks after feeding trial before unilateral eyestalk ablation (T1), and 1, 4, and 9 days after unilateral eyestalk ablation (T2, T3 and T4, respectively; Fig. 1A). All ovary samples were quickly frozen in liquid nitrogen and stored at −80 °C until use. The gonadosomatic index (GSI) of each shrimp was calculated as a percentage of ovary weight to body weight, classifying different ovarian developmental stages as the following: previtellogenic (stage I, GSI < 2), vitellogenic (stage 2, GSI = 2–4), early cortical rod stage (stage 3, GSI =<4–6) and mature stage (stage 4, GSI > 6)53. Differences among GSI from time points of the same feed group were considered significant Duncan; ANOVA (95% confidence).
RNA extraction was performed according a published protocol54. Briefly, frozen ovary tissues were pulverized in liquid nitrogen for RNA extraction using TRI-REAGENT (Molecular Research Center, USA). Contaminating genomic DNA was removed by treatment with DNase I at 0.15 U/µg total RNA at 37 °C for 30 min. The quality and quantity of the RNA were assessed using gel electrophoresis and a NanoDrop (ND-8000) spectrophotometer before subsequent experiments.
Transcriptome sequencing
To perform Iso-seq sequencing, we followed the protocol reported in Pootakham et al.55. Equal amounts of RNA samples from all treatments were pooled (Supplementary Table S1 indicates the samples that were used for Iso-seq), and poly(A) mRNAs were enriched using the Dynabeads mRNA Direct Purification Kit (Thermo Fisher Scientific, Waltham, USA). The RNA integrity was assessed using the Fragment Analyzer (Agilent Technologies, Santa Clara, USA) prior to the construction of the Iso-seq library. We followed the Iso-seq protocol from Pacific Biosciences, using the SMARTer PCR cDNA Synthesis Kit (Clontech, Mountain View, USA) to construct the library and the BluePippin Size Selection System (Sage Science, Beverly, USA) to partition the library into 1–2 kb, 2–3 kb and 3–6 kb ranges. The sequencing was performed on a PacBio RS II instrument using P4-C6 polymerase and chemistry with 360 min movie times (Pacific Biosciences, Menlo Park, USA).
To obtain short-read RNA sequences, Illumina sequencing was performed at Macrogen (South Korea). Each library was constructed from 1 μg total RNA from the ovaries of 2–4 individuals per treatment (Supplementary Table S1 indicates the samples that were used for RNAseq) using the TruSeq stranded Library Preparation kit (Illumina, San Diego, USA) and sequenced using Illumina NovaSeq™ 6000 instrument with 150 bp pair end read. Sequence data were deposited in the NCBI GenBank under the BioProject ID PRJNA542029.
Transcriptome assembly and annotation
Transcriptome assembly based on the Iso-seq data was performed according to the method reported in Pootakham et al.55. The PacBio SMRT Analysis Package (version 2.3; default parameters) was used to process raw reads from the PacBio RS II into error-corrected reads of insert. Adapter sequences, poly-A tails, artificial concatemers and 3′ truncated transcript sequences were removed using the Iso-seq protocol with default parameters. An additional round of error correction was carried out on the full-length non-chimeric transcripts using the PacBio ICE software without the Quiver step56. If the consensus accuracy was ≥ 0.99 (default parameter; https://github.com/ben-lerch/IsoSeq-3.0/blob/master/README.md), the polished isoforms were classified as “high-quality.”
Illumina short reads were quality-filtered and adapter-trimmed using TrimGalore (https://github.com/FelixKrueger/TrimGalore). FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to check data quality before and after trimming. After the removal of low-quality reads, an Illumina-based de novo transcriptome assembly was performed using Trinity v2.2.0 using the default parameter setting17. In order to obtain a complete ovarian transcriptome, we combined high-quality, full-length transcripts from the Iso-seq experiment with Trinity-assembled transcripts. To remove redundant sequences and obtain reference transcripts from the combined set of transcripts, we clustered these transcript sequences at 95% identity using UCLUST57. This reference transcriptome assembly was assigned function using BLASTX against custom NCBI non-redundant eukaryote protein database with an E-value cutoff of 1 × 10−10. BLASTX results were imported into Blast2Go Pro (version 5.2.4) for mapping and retrieving gene ontology (GO) terms associated with predicted sequences. The software also assigned the enzyme commission (EC) number and Kyoto encyclopedia of genes and genomes (KEGG) pathway annotations to the query sequences. The completeness of the transcriptome was evaluated using the coverage of the benchmarking universal single-copy orthologs (BUSCO)18.
Mapping and differential gene expression analysis
The pair-end Illumina reads were aligned to the reference transcriptome using Bowtie 2 version 2.2.958. The total mapped reads in each bam file were counted by HTSeq package through the HThtseq-count union function [14].
The R package DESeq. 2 [15] provides a test for differential expression using negative binomial generalized linear models, will operate to identify the differently expressed genes between shrimp fed polychaetes or pellet at different time points after eyestalk ablation. After read count normalization, log2 fold changes between shrimp fed polychaetes or pellet after eyestalk ablation will be calculated. Significant differently expressed genes will be identified with Bonferroni adjusted p-value of <0.05. Genes displayed in Fig. 4 were selected based on their expression profiles. Our ovarian transcriptome assembly contained a total of 11,694 non-redundant features. We first selected genes that were significantly (p-value < 0.05) up- or down-regulated during at least one time point (1,559 features). We subsequently clustered them based on their expression patterns and selected groups of genes that were affected by feed effect and eyestalk ablation using Cluster 3.0 software59. A total of 442 features exhibited changes in their expression level > 1.5 folds (Supplementary Table S5), and selected features with known functions were plotted in groups according to their similar expression patterns based on the results obtained from Cluster 3.0 software. Treeview version 1.6 was used to visualize gene expression data in Fig. 459. KEGG Automatic Annotation Server (KAAS) program was used to associate nucleotide sequences from 442 features to KEGG (Kyoto Encyclopaedia of Genes and Genomes) Orthology database (http://www.genome.jp/kegg/kaas/). Subsequently, the KEGGREST in R package was used to map KEGG Orthology (KO) to the pathways (Tenenbaum D (2019). KEGGREST: Client-side REST access to KEGG. R package version 1.26.1).
Quantitative real-time PCR (qPCR) validation of selected genes
Among differentially expressed genes, nine genes were selected for qPCR validation. Primers were designed using Primer Premier Program (Table 2). We followed the previously published protocol16. Briefly, 15 μg of total RNA were treated with RQ1-RNase-free DNase (Promega, USA) to eliminate contamination of genomic DNA. The first strand cDNA were synthesized from 1.5 µg of DNase-treated total RNA using an ImProm- IITM Reverse Transcription System Kit (Promega, USA) according to manufacturer’s instruction. Concentration and quality of the first strand cDNA were examined by Nanodrop UV (OD260/OD280). The cDNA of individual ovaries of black tiger shrimp from different time points were used as template. Each reaction was performed in a total volume of 20 µL containing 2x iQ™SYBR® Green Supermix (Bio-RAD, USA), 0.2 µg of cDNA template, 0.2 µM of primer pair. Cycling parameters were 95 °C for 2 min 30 s, followed by 40 cycles of 95 °C for 30 s, 58 °C for 20 s and 72 °C for 30 s. To ensure specificity of PCR products, melting curve analysis was performed from 55 °C to 95 °C with continuous fluorescence reading every 0.5 °C increment. The housekeeping gene Elongation factor 1 alpha (EF-1α) was used as an internal control60. All qPCRs were performed in three biological replications (n = 3).
For construction of the standard curve for each transcript, a plasmid containing the transcript was constructed by cloning the PCR product of the transcript into a pGEM-T easy vector (Promega, USA). The resulting vector was transformed into E. coli JM109. The plasmid was extracted and used as the template for construction of the standard curve by 10-fold serial dilutions (103–108 copy numbers).
Expression levels between feed groups were statistically tested by t-test (p < 0.05), and those among different time points post ablation of the same feed group were statistically tested by ANOVA followed by Duncan method (p < 0.05).
Change history
20 March 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Browdy, C. L. Recent developments in penaeid broodstock and seed production technologies: improving the outlook for superior captive stocks. Aquaculture 164, 3–21 (1998).
Preechaphol, R., Klinbunga, S., Yamano, K. & Menasveta, P. Molecular cloning and expression of progestin membrane receptor component 1 (Pgmrc1) of the giant tiger shrimp Penaeus monodon. Gen. and Comp. Endocrinol. 168, 440–449 (2010).
Leelatanawit, R. et al. Effects of polychaetes (Perinereis nuntia) on sperm performance of the domesticated black tiger shrimp (Penaeus monodon). Aquaculture 433, 266–275 (2014).
Lytle, J. S., Lytle, T. F. & Ogle, J. T. Polyunsaturated fatty acid profiles as a comparative tool in assessing maturation diets of Penaeus vannamei. Aquaculture 89, 287–299 (1990).
Meunpol, O., Iam-Pai, S., Suthikrai, W. & Piyatiratitivorakul, S. Identification of progesterone and 17α-hydroxyprogesterone in polychaetes (Perinereis sp.) and the effects of hormone extracts on penaeid oocyte development in vitro. Aquaculture 270, 485–492 (2007).
Wouters, R., Molina, C., Lavens, P. & Calderon, J. Lipid composition and vitamin content of wild female Litopenaeus vannamei in different stages of sexual maturation. Aquaculture 198, 307–323 (2001).
Sui, L. Y. et al. Effect of dietary HUFA on tissue fatty acid composition and reproductive performance of Chinese mitten crab Eriocheir sinensis (H. Milne-Edwards) broodstock. Aquacult Int. 19, 269–282 (2011).
Alava, V. R., Quinitio, E. T., de Pedro, J. B., Orosco, Z. G. A. & Wille, M. Reproductive performance, lipids and fatty acids of mud crab Scylla serrata (Forsskal) fed dietary lipid levels. Aquac. Res. 38, 1442–1451 (2007).
Huang, J. H., Jiang, S. G., Lin, H. Z., Zhou, F. L. & Ye, L. Effects of dietary highly unsaturated fatty acids and astaxanthin on the fecundity and lipid content of pond-reared Penaeus monodon (Fabricius) broodstock. Aquac. Res. 39, 240–251 (2008).
Aquacop. Penaied reared brood stock. closing the cycle of P. monodon, P. stylirostris and P. vannamei. Proc. of the World Mariculture Soc. 10, 445–452 (1979).
Kuo, C.-M., Chen, Y.-N., Fan, H.-F., Chuang, H.-C. & Hsieh, S.-L. Enhancement of vitellogenin synthesis by serotonin in the giant freshwater prawn Macrobrachium rosenbergii (de Man). Zool. Stud. 48, 597–606 (2009).
Guan, Z. B. et al. Participation of calmodulin in ovarian maturation induced by eyestalk ablation in red swamp crayfish Procambarus clarkii. Aquaculture Res. 44, 1625–1631 (2013).
Treerattrakool, S., Boonchoy, C., Urtgam, S., Panyim, S. & Udomkit, A. Functional characterization of recombinant gonad-inhibiting hormone (GIH) and implication of antibody neutralization on induction of ovarian maturation in marine shrimp. Aquaculture 428, 166–173 (2014).
Treerattrakool, S., Panyim, S. & Udomkit, A. Induction of Ovarian Maturation and Spawning in Penaeus monodon Broodstock by Double-Stranded RNA. Mar Biotechnol 13 (2011).
Hoang, T., Lee, S. Y., Keenan, C. P. & Marsden, G. E. Ovarian maturation of the banana prawn, Penaeus merguiensis de Man under different light intensities. Aquaculture 208, 159–168 (2002).
Uawisetwathana, U. et al. Insights into Eyestalk Ablation Mechanism to Induce Ovarian Maturation in the Black Tiger Shrimp. Plos One 6 (2011).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinforma. 31, 3210–3212 (2015).
Uengwetwanit, T. et al. Transcriptome-based discovery of pathways and genes related to reproduction of the black tiger shrimp (Penaeus monodon). Mar. Genomics 37, 69–73 (2018).
Nguyen, C. et al. De novo assembly and transcriptome characterization of major growth-related genes in various tissues of Penaeus monodon. Aquaculture 464, 545–553 (2016).
Bray, W. A., Lawrence, A. L. & Lester, L. J. Reproduction of Eyestalk-Ablated Penaeus stylirostris Fed Various Levels of Total Dietary Lipid. J. of the World Aquaculture Soc. 21, 41–52 (1990).
Perez-Velazquez, M., Lawrence, A. L., Gatlin, D. M., Gonzalez-Felix, M. L. & Bray, W. A. Replacement of fresh dietary components by a dry feed for successful maturation of male Litopenaeus vannamei (Boone) broodstock. Aquac. Res. 33, 1091–1095 (2002).
Wouters, R., Lavens, P., Nieto, J. & Sorgeloos, P. Penaeid shrimp broodstock nutrition: an updated review on research and development. Aquaculture 202, 1–21 (2001).
Avarre, J. C., Lubzens, E. & Babin, P. J. Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. Bmc Evol Biol 7 (2007).
Meusy, J. J. Vitellogenin, the extraovarian precursor of the protein yolk in Crustacea: A review. Reprod. Nutr. Dev. 20, 1–21 (1980).
Scott Quackenbush, L. Yolk synthesis in the marine shrimp, Penaeus vannamei. Comp. Biochem. and Physiol. Part. A: Physiol. 103, 711–714 (1992).
Tsukimura, B. Crustacean vitellogenesis: Its role in oocyte development. Am. Zool. 41, 465–476 (2001).
Tiu, S. H. K., Hui, J. H. L., Mak, A. S. C., He, J. G. & Chan, S. M. Equal contribution of hepatopancreas and ovary to the production of vitellogenin (PmVg1) transcripts in the tiger shrimp, Penaeus monodon. Aquaculture 254, 666–674 (2006).
Tiu, S. H. K., Chan, S.-M. & Benzie, J. From Hepatopancreas to Ovary: Molecular Characterization of a Shrimp Vitellogenin Receptor Involved in the Processing of Vitellogenin1. Biol. of Reprod. 79, 66–74 (2008).
Quackenbush, L. S. Yolk Synthesis in the Marine Shrimp, Penaeus vannamei1. Am. Zool. 41, 458–464 (2015).
Medeiros, M. N. et al. The role of eicosanoids on Rhodnius heme-binding protein (RHBP) endocytosis by Rhodnius prolixus ovaries. Insect Biochem. and Mol. Biol. 32, 537–545 (2002).
Schlotz, N., Sorensen, J. G. & Martin-Creuzburg, D. The potential of dietary polyunsaturated fatty acids to modulate eicosanoid synthesis and reproduction in Daphnia magna: A gene expression approach. Comp. Biochem. Phys. A 162, 449–454 (2012).
Wimuttisuk, W. et al. Insights into the prostanoid pathway in the ovary development of the penaeid shrimp Penaeus monodon. PLoS One 8, e76934–e76934 (2013).
Heckmann, L.-H., Sibly, R. M., Timmermans, M. J. & Callaghan, A. Outlining eicosanoid biosynthesis in the crustacean Daphnia. Front. in Zool. 5, 11–11 (2008).
Sreenivasula Reddy, P., Ramachandra Reddy, P. & Purna Chandra Nagaraju, G. The synthesis and effects of prostaglandins on the ovary of the crab Oziotelphusa senex senex. Gen. and Comp. Endocrinol. 135, 35–41 (2004).
Berkholtz, C. B., Shea, L. D. & Woodruff, T. K. Extracellular matrix functions in follicle maturation. Semin. Reprod. Med. 24, 262–269 (2006).
Berkholtz, C. B., Lai, B. E., Woodruff, T. K. & Shea, L. D. Distribution of extracellular matrix proteins type I collagen, type IV collagen, fibronectin, and laminin in mouse folliculogenesis. Histochem. Cell. Biol. 126, 583–592 (2006).
Deenarn, P. et al. Dynamics of fatty acid regulatory genes during ovarian development in Penaeus monodon. Reprod. 156, 527–544 (2018).
Tseng, D. Y., Chen, Y. N., Kou, G. H., Lo, C. F. & Kuo, C. M. Hepatopancreas is the extraovarian site of vitellogenin synthesis in black tiger shrimp, Penaeus monodon. Comp. Biochem. Phys. A 129, 909–917 (2001).
Wouters, R., Piguave, X., Bastidas, L., Calderon, J. & Sorgeloos, P. Ovarian maturation and haemolymphatic vitellogenin concentration of Pacific white shrimp Litopenaeus vannamei (Boone) fed increasing levels of total dietary lipids and HUFA. Aquac. Res. 32, 573–582 (2001).
Wei, B. et al. Effects of dietary lipids on the hepatopancreas transcriptome of Chinese mitten crab (Eriocheir sinensis). PLoS One 12, e0182087 (2017).
Chen, K. et al. Evaluation of different lipid sources in diet of pacific white shrimp Litopenaeus vannamei at low salinity. Aquaculture Rep. 2, 163–168 (2015).
Wimuttisuk, W. et al. Differential regulation of the lipoxygenase pathway in shrimp hepatopancreases and ovaries during ovarian development in the black tiger shrimp Penaeus monodon. Biochem. Bioph Res. Co. 487, 396–402 (2017).
Yang, J., Liu, D., Jing, W., Dahms, H.-U. & Wang, L. Effects of cadmium on lipid storage and metabolism in the freshwater crab Sinopotamon henanense. Plos One 8 (2013).
Qiu, G. F., Yamano, K. & Unuma, T. Cathepsin C transcripts are differentially expressed in the final stages of oocyte maturation in kuruma prawn Marsupenaeus japonicus. Comp. Biochem. Phys. B 140, 171–181 (2005).
Baarends, W. M., Roest, H. P. & Grootegoed, J. A. The ubiquitin system in gametogenesis. Mol. and Cell. Endocrinol. 151, 5–16 (1999).
Grote, P. & Conradt, B. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C-elegans. Dev Cell 11 (2006).
Karoonuthaisiri, N. et al. ReproArray(GTS): A cDNA microarray for identification of reproduction-related genes in the giant tiger shrimp Penaeus monodon and characterization of a novel nuclear autoantigenic sperm protein (NASP) gene. Comp. Biochem. Phys. D. 4, 90–99 (2009).
Loredo-Ranjel, R., Fanjul-Moles, M. L. & Escamilla-Chimal, E. G. Crustacean hyperglycemic hormone is synthesized in the eyestalk and brain of the crayfish Procambarus clarkii. Plos One 12 (2017).
Udomkit, A., Treerattrakool, S. & Panyim, S. Crustacean hyperglycemic hormones of Penaeus monodon: cloning, production of active recombinant hormones and their expression in various shrimp tissues. J. Exp. Mar. Biol. Ecol. 298, 79–91 (2004).
Jimenez, G., Gonzalez-Reyes, A. & Casanova, J. Cell surface proteins Nasrat and Polehole stabilize the Torso-like extracellular determinant in Drosophila oogenesis. Gene Dev. 16, 913–918 (2002).
Klinbunga, S. et al. Molecular Cloning and Expression Analysis of Ovary-Specific Transcript 1 (Pm-OST1) of the Giant Tiger Shrimp, Penaeus monodon. Zool. Sci. 26, 783–790 (2009).
Preechaphol, R., Klinbunga, S., Yamano, K. & Menasveta, P. Molecular cloning and expression of progestin membrane receptor component 1 (Pgrnrc1) of the giant tiger shrimp Penaeus monodon. Gen. Comp. Endocr. 168, 440–449 (2010).
Leelatanawit, R. et al. Transcriptomic Analysis of Male Black Tiger Shrimp (Penaeus monodon) After Polychaete Feeding to Enhance Testicular Maturation. Mar. Biotechnol. 19, 125–135 (2017).
Pootakham, W. et al. Development of a Novel Reference Transcriptome for Scleractinian Coral Porites lutea Using Single-Molecule Long-Read Isoform Sequencing (Iso-Seq). Front Mar Sci 5, Unsp 122 (2018).
Gordon, S. P. et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS One 10, e0132628 (2015).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinforma. 26, 2460–2461 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998).
Leelatanawit, R., Klanchui, A., Uawisetwathana, U. & Karoonuthaisiri, N. Validation of Reference Genes for Real-Time PCR of Reproductive System in the Black Tiger Shrimp. Plos One 7 (2012).
Acknowledgements
We thank Dr. Rungnapa Leelatanawit, Ms. Somjai Wongtripop, Ms. Thaninthorn Raweeratanapong and SGIC members for the shrimp samples and facility to conduct the eyestalk ablation experiment. This project was funded by National Center for Genetic Engineering and Biotechnology (RI grant number P1851724 and Platform grant number P1651718), National Science and Technology Development Agency (grant number P1950419), and partially supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 734486.
Author information
Authors and Affiliations
Contributions
K.S., W.P. and N.K. conceived of the study. K.S., J.K., D.S. and T.Y. collected samples and performed laboratory work, library construction and sequencing. W.P., C.S., U.U. and I.N. performed bioinformatics analyses. K.S. and W.R. performed quantitative real-time PCR analysis. W.P., U.U., I.N. and N.K. wrote the manuscript and all authors reviewed it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sittikankaew, K., Pootakham, W., Sonthirod, C. et al. Transcriptome analyses reveal the synergistic effects of feeding and eyestalk ablation on ovarian maturation in black tiger shrimp. Sci Rep 10, 3239 (2020). https://doi.org/10.1038/s41598-020-60192-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60192-2
This article is cited by
-
RNA-Seq reveals divergent gene expression between larvae with contrasting trophic modes in the poecilogonous polychaete Boccardia wellingtonensis
Scientific Reports (2021)
-
Transcriptomic analysis of the black tiger shrimp (Penaeus monodon) reveals insights into immune development in their early life stages
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.