Abstract
DJ-1 is a multifunctional protein that plays roles in transcriptional regulation and antioxidative stress, and loss of its function is thought to result in the onset of Parkinson's disease (PD). Here, we report that DJ-1 was sumoylated on a lysine residue at amino-acid number 130 (K130) by PIASxα or PIASy. The K130 mutation abrogated all of the functions of DJ-1, including ras-dependent transformation, cell growth promotion and anti-UV-induced apoptosis activities. Sumoylation of DJ-1 was increased after UV irradiation concomitant with a pI shift to an acidic point of DJ-1. Furthermore, L166P, a mutant DJ-1 found in PD patients, and K130RX, an artificial mutant containing four mutations in DJ-1, were improperly sumoylated, and they became insoluble, partly localized in the mitochondria and degraded by the proteasome system. Both L166P-expressing cells and DJ-1-knockdown cells were found to be highly susceptible to UV-induced cell apoptosis.
Similar content being viewed by others
Introduction
DJ-1 was first identified by our group as a novel candidate of the oncogene product that transformed mouse NIH3T3 cells in cooperation with activated ras.1 The human DJ-1 gene is mapped at chromosome 1p36.2–p36.3, where a hot spot of chromosome abnormalities has been reported in several tumors.2 However, the mechanism by which cells are transformed has not been clarified. Another group has also identified RS, another name for human DJ-1, as a regulatory component of an RNA-binding protein complex.3 Furthermore, CAP-1 or SP22, a rat homologue of human DJ-1, has been identified by other laboratories as a key protein related to infertility of male rats that were exposed to sperm toxicants such as ornidazole and epichlorohydrin. DJ-1 in the sperm and epididymis decreased in parallel with the subsequent infertility of the rats and was later found to be related to infertility.4, 5, 6, 7, 8, 9 It has also been shown that DJ-1 is a circulating tumor antigen in breast cancer, in which DJ-1 is secreted from cells to serum,10 and that DJ-1 is overexpressed in smoker-derived lung adenocarcinoma.11 DJ1 is therefore thought to play roles in both somatic cells and sperm.
We have shown that DJ-1 acts as a positive regulator for the androgen receptor (AR) by sequestering PIASxα or DJBP.12, 13, 14 PIASxα, a member of the PIAS family of proteins, and DJBP, a novel DJ-1-binding protein, function as negative regulators for AR by preventing AR DNA-binding activity and by recruiting histone deacetylase complex, respectively.12, 13 We have also showed that DJ-1 bound to p53 to restore p53 transcription activity inhibited by Topors/p53BP3 and that transforming activity of DJ-1 was stimulated by Abstrakt, another DJ-1-binding protein.15, 16
Deletion and point (L166P) mutations of DJ-1 have recently been shown to be responsible for the onset of familial Parkinson's disease (PD), PARK7,17 and other homozygous and heterozygous mutations of DJ-1 have been identified in patients with familial or sporadic PD.18, 19, 20 PD involves an irreversible degeneration of the dopaminergic nigrostriatal pathway. Genes responsible for rare familial early-onset PD, including α-synuclein,21 Parkin22 and UCH-L1,23 have been identified, and they are thought to play a role in ubiquitin–proteasome dysfunction in PD. L166P DJ-1 has been reported to be degraded, in part, by the ubiquitin–proteasome system.24, 25, 26, 27, 28 DJ-1 was shown to be expressed in almost all brain tissues in healthy men and patients with neurodegenerative diseases, including PD, Pick's disease and multiple system atrophy.29, 30, 31 Various lines of evidence also suggest that oxidative stresses contribute to the cascade leading to dopaminergic cell degeneration in PD,32, 33, 34, 35 and expression of DJ-1 was shown to be induced by oxidative stresses.36, 37, 38, 39 A pI shift of DJ-1 towards a more acidic isoform has been observed in PD patients.29 We previously reported that DJ-1 plays a role in the antioxidative stress reaction, in which reactive oxygen species (ROS) were eliminated in vitro and in vivo by oxidizing DJ-1 itself, and that mutations of DJ-1, including L166P, lead to cell death.40, 41 Recently, PINK1, a causative gene product of PARK6, has also been suggested to play a role in an antioxidative stress response,42 but precise roles of PINK1 are not clarified.
SUMO-1 conjugation to lysine residues of proteins is catalyzed in three steps by E1, E2 and E3 enzymes and modulates interactions, localization, activity and stability of proteins to be sumoylated (see recent reviews and references therein).43, 44 E3 SUMO-1 ligases promote attachment of the SUMO modifier to target proteins, and some SUMO-1 ligases have been identified. PIAS family proteins, which include PIAS1, PIAS3, PIASxα, PIASxββ and PIASy, were initially identified as negative regulators of STAT-transcription factors and later found to function as transcriptional coregulators in various other cellular pathways, including Wnt signaling, the p53 pathway and steroid hormone signaling. PIAS proteins were then found to act as E3 SUMO-1 ligases that stimulate SUMO-1 conjugation to proteins, including p53,45, 46, 47 AR,47 Jun46 and Lef-1.48 In most cases, the PIAS/SUMO pathway appears to be an important mechanism of transcriptional regulation.
In this study, we found that mutation of lysine at amino-acid number 130 of DJ-1 abrogated all of the functions of DJ-1 and that improperly sumoylation to L166P mutant DJ-1 makes DJ-1 insoluble, which may trigger the onset of PD.
Results
SUMO-1 conjugation of DJ-1 in cells
We have reported that DJ-1 binds to PIASxα and regulates the AR transcription activity.12 In addition to PIASxα, both Ubc9 and SUMO-1 were identified as DJ-1-binding proteins by a yeast two-hybrid screening. Ubc9 is an E2 enzyme in the SUMO-1 conjugation system. These findings indicate the possibility of sumoylation of DJ-1. To test this possibility, human H1299 cells were lysed in SDS-containing buffer, boiled, and immunoprecipitated with an anti-DJ-1 antibody or nonspecific IgG. Precipitates were analyzed by Western blotting with an anti-DJ-1 antibody or an anti-SUMO-1 antibody (Figure 1). In addition to a 26-kDa band corresponding to DJ-1, a 44 kDa band appeared in the immunoprecipitate with the anti-DJ-1 antibody but not with IgG (Figure 1, lanes 2 and 1, respectively). Since SUMO-1 conjugation to proteins is known to give an addition of approximately 20 kDa to proteins in SDS-polyacrylamide gel and since the 44-kDa band reacted with the anti-SUMO-1 antibody, the results clearly showed that DJ-1 was sumoylated in vivo in H1299 cells (Figure 1, lane 4).
Then EST clones for cDNAs encoding DJ-1 homologs of chicken, fish (Atlantic salmon) and Drosophila were obtained, and monkey DJ-1 cDNA was constructed by RT-PCR using total RNA from monkey COSI cells as a template. A search for amino-acid sequences for DJ-1 homologs of Xenopus, nematode and Drosophila were carried out in the NIH-NCI Database. Interestingly, two homologs of DJ-1 were found to be present both in nematode and Drosophila. Amino-acid sequences of DJ-1s were found to be highly conserved among species, and the identities of the sequences of monkey, mouse, rat, chicken, Xenopus, fish and Drosophila to that of human DJ-1 are 98, 91, 91, 89, 78, 80, 52 (nematode 1), 44 (nematode 2), 52 (Drosophila 1) and 58% (Drosophila 2), respectively (Figure 2a). Of 16 lysines, candidates for the sumoylated amino acid in human DJ-1, the lysine at amino-acid number 130 was found to be conserved among all of the species (Figure 2a). We therefore converted all of the lysines of human DJ-1 to arginine and tested the SUMO-1 conjugation to these mutants of DJ1s in H1299 cells into which FLAG-DJ-1, HA-Ubc9 and T7-SUMO-1 were cotransfected. At 48 h after transfection, the cells were lysed in SDS-containing buffer, boiled, and immunoprecipitated with an anti-FLAG antibody. Precipitates were analyzed by Western blotting with the anti-FLAG antibody (Figure 2b). The results showed that wild-type DJ-1 and all of the arginine-substituted mutants except for K130R were sumoylated, indicating that K130 is a major sumoylation site of DJ-1 (Figure 2, lane 12). It was noted that both the K99R mutant and its sumoylated form of DJ-1 run slowly in gel, suggesting that this mutant possesses a conformation different from that of wild-type DJ-1 (Figure 2, lane 10).
Alignment of deduced amino-acid sequences of DJ-1s from human to Drosophila and identification of the sumoylation site on DJ-1. (a) Accession numbers for amino-acid sequences are D61380 (human), AB073863(monkey), AB015652 (mouse), AJ007291 and AF157511 (rat), AB076264 (chicken), AF394958 (Xenopus), AB079598 (Atlantic salmon), AAB37889 (nematode 1), AAK18904 (nematode 2), AB079599 (Drosophila 1) and AAF58316 (Drosophila 2). The identical amino acids are drawn in gray, and the lysines are indicated by dots. Lysines corresponding to lysine at amino-acid number 130 of human DJ-1 are boxed. (b) H1299 cells were transfected with FLAG-DJ-1 and lysine-substituted mutants ofDJ-1. At 48 h after transfection, proteins extracted from transfected cells were immunoprecipitated with an anti-FLAG antibody and blotted with the anti-FLAG antibody as described in Materials and Methods
We previously reported that DJ-1 binds to both PIASxα and PIASy,12 both of which are known to function as an E3 SUMO-1 ligase. We therefore tested whether PIASxα and PIASy are E3 SUMO-1 ligases for DJ-1. H1299 cells were cotransfected with FLAG-DJ-1, HA-Ubc9, T7-SUMO-1 and various amounts of HA-PIASxα or -PIASy and SUMO-1 conjugation to DJ-1 was examined. Expressions of HA-PIASxα and -PIASy were first confirmed by blotting the precipitates with an anti-HA antibody (Figure 3a, lanes 4–9, 11 and 12 in lower part). The results showed that while the HA-tag alone did not stimulate sumoylation of DJ-1, both HA-PIASxα and -PIASy stimulated sumoylation of wild-type DJ-1 but not K130R in a dose-dependent manner (Figure 3a), indicating that both PIASxα and PIASy work as E3 SUMO-1 ligases for DJ-1. To examine the effect of PIASxα on sumoylation to endogenous DJ-1, H1299 cells were cotransfected with PIASxα-HA, T7-SUMO-1 and Ubc9-HA, and SUMO-1 conjugation to DJ-1 was examined. As in the case of the ectopic expression system shown in Figure 3a, endogenous DJ-1 was sumoylated by PIASxα in a dose-dependent manner (Figure 3b, lanes 2–5), and similar results were obtained when PIASy was cotransfected into H1299 cells (data not shown). These results clearly indicate that both PIASxα and PIASy are E3 SUMO-1 ligases for DJ-1.
Stimulation of SUMO-1 conjugation to DJ-1 by PIASxα and PIASy. (a) H1299 cells were cotransfected with FLAG-DJ-1, T7-SUMO-1, Ubc9-HA and various amounts of PIASxα-HA or PIASy. At 48 h after transfection, proteins extracted from transfected cells were immunoprecipitated with an anti-FLAG antibody and blotted with the anti-FLAG antibody or an anti-HA antibody as described in Materials and Methods. The amounts of expression vectors for PIASxα-HA and PIASy were 2.5, 5.0 and 7.5 μg in lanes 4 and 7, 5 and 8, and 6 and 9, respectively. (b) H1299 cells were cotransfected with T7-SUMO-1, Ubc9-HA and various amounts of PIASxα-HA. At 48 h after transfection, proteins extracted from transfected cells were immunoprecipitated with an anti-DJ-1 antibody and analyzed by Western blotting with the anti-DJ-1 antibody or an anti-SUMO-1 antibody as described in Materials and Methods. The amounts of expression vectors for PIASxα-HA were 2.5, 5.0 and 7.5 μg in lanes 3, 4 and 5, respectively
Abrogation of DJ-1 activities by K130 mutation
To know the importance of K130 for DJ-1 activities, transformation activities of several lysine-substitution mutants of DJ-1 were tested in rat 3Y1 cells by cotransfection of DJ-1 mutants with activated ras (Figure 4a). As previously reported in mouse NIH3T3 cells,1 wild-type DJ-1 gave a number of transformed foci only in the case of cotransfection with activated ras. While DJ-1 mutants of both K62, 63R, in which lysines at #62 and 63 were changed to arginine, and K93R still gave numbers of foci similar to that given by wild-type DJ-1, DJ-1 mutants of K130R and K130RX, in which three other amino acids, S57R, E96G and H126Y, had been changed,12 as well as K130 gave smaller numbers of foci than that without activated ras. These results clearly indicate that K130 in DJ-1 is of functional importance for the transforming activity of DJ-1.
Effects of lysine mutations of DJ-1 on the transforming and cell growth promoting activities of DJ-1. (a) Rat 3Y1 cells were transfected with expression vectors for activated wild-type DJ-1or mutants of DJ-1 with or without activated H-ras. In total, 14 days after transfection, the transformed cell foci were counted. (b) Rat 3Y1 cells were transfected with neomycin-resistant gene-containing expressionvectors for wild-type DJ-1 or mutants of DJ-1, and they were cultured in the presence of G418. At 14 days after transfection, the G418-resistant cell colonies were counted. Pictures of the colonies are shown
Many proteins possessing transforming activity often have cell growth-stimulating activity. To test this activity of DJ-1, rat 3Y1 cells were transfected with the expression vectors for wild-type and mutant forms of DJ-1 that were cloned in pcDNA3 containing a neomycin-resistant gene, and they were cultured in a medium containing G418. G418-resistant colonies were then counted 14 days after transfection (Figure 4b). Transfection with c-Myc as a positive control gave a large number of colonies, while transfection with the vector alone or no-DNA control (none) gave few or no colonies. Wild-type or non-K130 mutants of DJ-1, K62, 63R and K93R, gave similar numbers of colonies to those of c-Myc. K130R and K130RX mutants of DJ-1, on the other hand, abolished the colony-forming activity to the level of the vector alone. These results indicate that DJ-1 possesses cell growth-promoting activity that is dependent upon K130.
Induction of DJ-1 expression and stimulation of SUMO-1 conjugation to DJ-1 by UV irradiation
Expression of DJ-1 was induced in various cells that had been subjected to treatment with paraquat, endotoxin or iron, which yielded ROS.36, 37, 38, 39 We also showed that expression of DJ-1 was induced in human neuroblastoma SH-SY5Y cells treated with hydrogen peroxide.41 Since ultraviolet (UV) irradiation is known also to induce ROS, we first examined whether DJ-1 expression is induced after 20 J/m2 UV irradiation by using p53-positive and p53-negative cell lines, ME180 and H1299 cells, respectively, in which p53 is a positive control as an induced protein against UV irradiation (Figure 5A). In ME180 cells, while expression of p53 was induced at 2 h after UV irradiation, expression of DJ-1 was induced at 15 min after UV irradiation and the level peaked at 30–45 min and then gradually decreased up to 3 h after UV irradiation (Figure 5A-a). In H1299 cells, induction of DJ-1 expression started at 15 min after UV irradiation and the level peaked at 30–45 min and then gradually decreased until 6 h after UV irradiation (Figure 5A-b). These results suggest that expression of DJ-1 is induced by UV irradiation in a p53-independent manner.
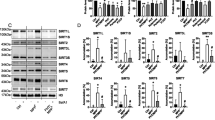
Induction of expression, pI shift and stimulation of sumoylation of DJ-1. (A) ME180 cells (a) and H1299 cells (b) were irradiated with 20 J/m2 of UV, and at various times after irradiation, proteins extracted from cells were blotted with an anti-p53 (FL-393, Santa Cruz), anti-DJ-1 polyclonal and anti-actin antibodies (upper panel). Bands that reacted with the antibody were quantitated using a densitometer (lower panel). (B) H1299 cells were irradiated with 20 and 50 J/m2 of UV, and proteins in the extracts were then analyzed by isoelectric focusing phoresis gel as described in Materials and Methods. (C) H1299 cells were irradiated with 20 J/m2 of UV. At various times after irradiation, proteins extracted from cells were immunoprecipitated with an anti-DJ-1 polyclonal antibody and blotted with an anti-DJ-1 monoclonal antibody or anti-actin antibody as described in Materials and Methods. (D) ME180 cells were irradiated with 20 J/m2 of UV. At various times after irradiation, cells were stained with an anti-DJ-1 polyclonal antibody. The cells were then reacted with an FITC-conjugated anti-rabbit IgG and observed under a confocal laser fluorescent microscope. The cells were also stained with DAPI and MitoTracker-red (Molecular probe)
Shifts of the isoelectric point of DJ-1 in cells caused by oxidative stresses have been reported.36, 37, 38, 39, 40, 41 H1299 cells were irradiated with 20 and 50 J/m2 UV, and proteins prepared from cells were separated on isoelectric focusing gel over the pI range of 5–8 (Figure 5B). After blotting with an anti-DJ-1 antibody, pI of DJ-1 was found to be shifted to a more acidic point, from 6.3 to 5.2, at 1 h after UV irradiation (Figure 5B). SUMO-1 conjugation to DJ-1 was also found to be stimulated at 25 min, peak at 30 min and then gradually decreased after UV irradiation (Figure 5C). These results suggest that DJ-1 is first sumoylated and then oxidized after UV irradiation.
ME180 cells after UV irradiation were stained with a rabbit anti-DJ-1 polyclonal antibody, reacted with an FITC-conjugated anti-rabbit IgG, and visualized under a confocal laser microscope (Figure 5D). Nuclei and the mitochondria in the cytoplasms were also stained with DAPI and MitoTracker, respectively. Endogenous DJ-1 was found to be localized mainly in the cytoplasm and sparsely in the nucleus without UV irradiation (Figure 5D, time 0 h). After UV irradiation, almost all the DJ-1 were translocated to the nucleus, which was shown in the merged figures as light blue color (Figure 5D, time 0.25–0.75 h). At 1 h after UV irradiation, some DJ-1 were relocalized in the cytoplasm and almost all the DJ-1 were located in the cytoplasm at 24 h after UV irradiation. These results indicate that a portion of DJ-1 is shuttled in between the cytoplasm and nucleus of UV-irradiated H1299 cells.
Improper sumoylation and insoluble form of L166P mutant of DJ-1 found in PD patients
Homozygous point mutations of the DJ-1 gene have been found in Italian kindred of PD patients,17 and its gene product L166P mutant form of DJ-1 has been reported to be unstable and to be degraded, in part, by the ubiquitin–proteasome system.24, 25, 26, 27, 28, 49 We therefore examined the sumoylation status of L166P DJ-1 (Figure 6). H1299 cells were transfected with FLAG-tagged wild type and three mutants of DJ-1 together with SUMO-1 and Ubc9, and protein extracts from transfected cells were immunoprecipitated with an anti-FLAG antibody. Precipitates were then separated on two gels and analyzed by Western blotting with the anti-FLAG antibody or an anti-SUMO-1 antibody (Figure 6A). Sumoylation of wild-type DJ-1 but not K130R DJ-1 was first confirmed (Figure 6A-a, lanes 1, 2, 6 and 7). Characteristics of the band of 62 kDa observed above the DJ-1-SUMO-1 bands are not clear at present. It is interesting that L166P and K130RX showed ladder bands over the range of SUMO-1-DJ-1 in a blot with the anti-FLAG antibody and that these bands also reacted with the anti-SUMO-1 antibody (Figure 6A-a, lanes 8, 9, 3 and 4), indicating that L166P and K130RX mutants of DJ-1 are conjugated with multi- or poly-SUMO-1, or alternatively that these mutant proteins became misfolded, resulting in the formation of the abnormal aggregates. Since a lysine residue at amino-acid number 130 (K130) in K130RX DJ-1 was changed to arginine, it is possible that sumoylation occurs on lysine residue(s) other than K130 in K130RX DJ-1 and that this is also true for sumoylation of L166P DJ-1. To examine this possibility, K130 in L166P DJ-1 was changed to arginine and sumoylation assay of this mutant (K130R, L166P DJ-1) was carried out. This mutant showed ladder bands as did K130RX and L166P (Figure 6A-a, lanes 5 and 10), indicating that sumoylation occurred some lysine residues, including L130, in L166P DJ-1. This improper sumoylation of L166P and K130RX mutants of DJ-1 was also observed in an in vitro reconstituted system using recombinant proteins comprised of SAE1/SAE2 complex, Ubc9 and SUMO-1 (Figure 6A-b, lanes 3 and 4), suggesting that the structure of L166P and K130RX mutants of DJ-1 possess easier accessibility to be sumoylated than does wild-type DJ-1 and was changed to be improperly sumoylated.
Improper sumoylation of L166P mutant of DJ-1. (A (a)) H1299 cells were cotransfected with FLAG-DJ-1 or mutants, T7-SUMO-1 andUbc9-HA. At 48 h after transfection, proteins extracted from transfected cells were immunoprecipitated with an anti-FLAG antibody and analyzed by Western blotting with the anti-FLAG antibody or an anti-SUMO-1 antibody as described in Materials and Methods. (b) In vitro sumoylation of DJ-1 and its mutant were carried out using the reaction mixture containing 35S-DJ-1 and its mutated DJ-1 and the recombinant proteins of E1 enzyme, Ubc9 and SUMO-1 as described in Materials and Methods. Labeled proteins in the mixture were separated on polyacrylamide gel and visualized by fluorography. (B) 293T cells were transfected with FLAG-DJ-1 or its mutants. At 48 h after transfection, soluble proteins was first extracted from transfected cells with 0.1% SDS-containing buffer and insoluble proteins were then dissolved with 2% SDS. These were analyzed by Western blotting with an anti-FLAG antibody
We then examined the solubility of wild-type DJ-1 and mutants of DJ-1 in cells. Human 293T cells were transfected with FLAG-tagged wild-type DJ-1 and four mutants of DJ-1, L166P, K130R, K130RX and C106S. An amino acid at C106 is a putative active center for protease activity of DJ-1, and a C106S mutant had no protease and antioxidative stress activities.28, 41, 50, 51, 52 At 48 h after transfection, the soluble proteins were extracted with 0.1% NP-40 and insoluble proteins were further dissolved in 2.0% SDS, and these proteins were analyzed by Western blotting with an anti-FLAG antibody (Figure 6B). While wild-type DJ-1, C106S DJ-1 and K130R DJ-1 were in soluble forms, both L166P and K130RX were found to be in insoluble forms. Since insoluble forms of DJ-1 have been observed in patients with degenerative neuronal diseases, it is interesting that improperly sumoylated DJ-1 becomes insoluble (see Discussion).
We then examined the cellular localization of these mutants of DJ-1. H1299 cells were transfected with FLAG-tagged wild-type DJ-1 and four mutants of DJ-1. At 48 h after transfection, cells were stained with an ant-FLAG antibody and visualized with an FITC-conjugated anti-mouse antibody (Figure 7). Since L166P DJ-1 has been reported to be localized, in part, in the mitochondria17 and we have reported that DJ-1 also played a role in antiendoplasmic reticulum (ER) stress-induced cell death,40 cells were also reacted with MitoTracker and an anti-GRP78 to see the mitochondria and ER, respectively (Figures 7a and data not shown, respectively). Nuclei were also stained with DAPI. Wild-type DJ-1 and K130R DJ-1 were localized both in the cytoplasm and nucleus, and some portion of DJ-1 were also localized in the mitochondria as shown by merged figures in which the green and red have become yellow (Figure 7a, k and l). K130RX, L166P DJ-1 and a double mutant (K130R, L166P) were, on the other hand, found to be predominantly localized in the cytoplasm and almost all of them were localized in the mitochondria (Figure 7a, m, n and o). Neither wild-type DJ-1 nor any of the four mutants of DJ-1, on the other hand, was found to be localized in the ER in cells without ER stress (data not shown). Wild-type DJ-1 and the four mutants of DJ-1 were also not found to be localized in the Golgi apparatus (data not shown).
Localization of DJ-1 and its mutants in cells. H1299 cells were transfected with FLAG-wild-type (Wt) or its mutants. At 48 h after transfection, cells were fixed, reacted with a combination of an anti-FLAG antibody (a–e), DAPI and MitoTracker (f–j), and visualized with a fluorescein isothiocyanate-conjugated anti-mouse antibody (a–e) and self-fluorescences of DAPI and MitoTracker (f–j). The two figures have been merged (k–o)
Since L166P DJ-1 has been reported to be degraded, in part, by the ubiquitin–proteasome system24, 25, 26, 27, 28, 49 and DJ-1 possess cysteine protease activity,28 we then examined the stability of mutants of DJ-1 against degradation by proteasome and cysteine protease using their specific inhibitors, MG132 and E64, respectively. Proteins were extracted from NIH3T3 cells transfected with FLAG-tagged wild-type DJ-1 and mutants of DJ-1 in the presence or absence of MG132 or E64 and blotted with an anti-FLAG antibody or an antiactin antibody (Figure 8). The results first showed that the amounts of L166P and K130RX DJ-1 were smaller than those of wild-type DJ1 and K130R DJ-1 in the absence of MG132 (Figure 8, lanes 1, 4, 7, 10, 13 and 16) and that E64 had no effect on the stability of wild-type DJ-1 or any of the mutants of DJ-1 (Figure 8, lanes 3, 6, 9, 12, 15 and 18). Although wild-type DJ-1 and K130R DJ-1 were not affected by MG132, the amounts of L166P and K130RX increased partially in the presence of MG132 (Figure 8, lanes 14 and 17 in the middle part), suggesting that K130RX and L166P DJ-1 were degraded, in part, by the proteasome system. It was noted that slow-migrating bands of K130RX DJ-1 appeared in the presence of MG132 (Figure 8, lane 17 in the upper part). Although it is possible that these bands correspond to the ubiquitinated K130RX DJ-1, it is not certain at present due to the inability of identification of these bands against an antiubiquitin antibody (data not shown). These results suggest that mutants of DJ-1 localized in the mitochondria are improperly sumoylated, insoluble and unstable.
Stability of DJ-1 and its mutants. Mouse NIH3T3 cell lines harboring FLAG-wild-type DJ-1 and its mutants were cultured in the presence of 10 mM MG132 or 8 μg/ml of E64 for 12 h. Proteins extracted from cells were analyzed by Western blotting with anti-FLAG and antiactin antibodies. Host indicates parental NIH3T3 cells
Loss of antiapoptosis activity of DJ-1 mutants
We then examined the effects of K130R and L166P mutations of DJ-1 on UV-induced apoptosis. Mouse NIH3T3-Wt, -K130R and -L166P cells, which are stable cell lines expressing wild-type, K130R and L166P DJ-1 as described previously,41 and NIH3T3-D2 cells, which are stable DJ-1-knockdown cells expressing siRNA toward DJ-1,49 were irradiated with two doses of UV. At 22 h after irradiation, DNA from cells was stained with propidium iodide and the distribution of cells in the cell cycle was analyzed by flow cytometry (Figure 9). DJ-1 expression in D2 cells decreased to about 10% of that of parental NIH3T3 cells (Figure 9a) and the characteristics of D2 cells has been described.49 Apoptotic cells that appeared at the position below the G1/G0 phase were counted using the ‘ModiFit LT’ program (Figure 9b). As in the case of hydrogen peroxide-induced cell death,41 cells harboring exogenously added wild-type DJ-1 were much more resistant to UV than were parental nontransfected cells (host). Of the cell lines harboring mutants, on the other hand, the cell line harboring L166P was found to be most sensitive to UV and the K130R line followed in terms of sensitivity to UV. DJ-1-knockdown cells were found to have similar sensitivity to that of L166P cells. These results clearly indicate that DJ-1 plays a role in protection against UV-induced apoptosis and that mutations or knockdown of DJ-1 lead to cell death.
Abrogation of antiapoptosis activity of DJ-1 by mutations. (a) Proteins extracted from NIH3T3 cells (host) and DJ-1-knockdown cells (D2) were analyzed by Western blotting with anti-mouse DJ-1 and antiactin antibodies. The proteins that reacted with primary antibodies were visualized with IRDye800conjugated or Alexa Fluor680-conjugated secondary antibodies using an infrared imaging system (Odyssey, LI-COR). (b) Mouse NIH3T3 cell lines harboring FLAG-wild-type DJ-1 and its mutants were irradiated with 100 and 2000 J/m2 UV. At 22 h after irradiation, DNA in cells was stained with propidium iodide and the distribution of cells in the cell cycle was analyzed by flow cytometry. Distribution of cells in the cell cycle by analysis of flow cytometry was quantitated by using a ‘ModiFit LT’ program. Results (n=3) were statistically processed using Student's t-test. Significance: *P<0.01 versus apoptotic cells in wt-DJ-1 cells
Discussion
SUMO-1 conjugation to DJ-1
In this study, we first found that a lysine at amino-acid number 130 of DJ-1 (K130) was sumoylated by PIASxα and PIASy in cells and that this sumoylation of DJ-1 was essential for DJ-1 to exert its full activities, including ras-dependent transforming and cell growth-stimulating activities. PIAS family proteins, which include PIAS1, PIAS3, PIASxα, PIASxββ and PIASy, are known to function as E3 SUMO-1 ligases and the proteins to be sumoylated by each PIAS family protein were identified.43, 44 In most case, transcription factors have been identified as target proteins for PIAS family proteins and DJ-1 is also a transcription factor as a coactivator to regulate expressions of genes for AR, p53 and PTEN.12, 13, 15, 53, 54 Since identifications of almost all the proteins that had been sumoylated were performed by overexpression system using expression vectors for SUMO-1 and Ubc9, the level of sumoylation of DJ-1, which was observed without overexpression of SUMO-1 or Ubc9, is thought to be high.
DJ-1 expression and its sumoylation were induced by UV irradiation and DJ-1 was translocated from the cytoplasm to nucleus. Since induction of DJ-1 expression after irradiation of UV occurred both in p53 (−) and p53 (+) cells and this occurred faster than did induction of p53 expression in p53 (+) cells, induction of DJ-1 expression by UV irradiation is p53-independent. UV irradiation is known to induce ROS and to give the oxidative stress to cells, and DJ-1 expression has been reported to be induced by several chemicals that had also been known to induce ROS.36, 37, 38, 39 UV irradiation of cells also shifted the pI of DJ-1 to acidic points, as in the case of direct addition of hydrogen peroxide, one of the ROS, to human SH-SY5Y and mouse NIH3T3 cells, in which ROS were scavenged by DJ-1, as described previously.41 The amount of pI-shifted DJ-1, however, was again reduced 4 h after UV irradiation concomitant with a reduction of the amount of the sumoylated form of DJ-1 and relocalization of DJ-1 in the cytoplasm, suggesting that self-oxidation of DJ-1 following SUMO-1 conjugation to DJ-1 occurs depending on the amounts of ROS in cells and that the sumoylated form of DJ-1 is an active form to eliminate ROS. High-dose of UV irradiation of cells induces apoptosis, and an NIH3T3 cell line expressing exogenously added wild-type DJ-1 became resistant to UV-induced apoptosis compared to the parental cells. This antiapoptotic activity of DJ-1 was abrogated by the mutation into an amino acid K130, a sumoylation site of DJ-1, as well as those of ras-dependent transforming and cell growth-stimulating activities, indicating that SUMO-1 conjugation to K130 of DJ-1 is essential for DJ-1 to function.
Improper SUMO-1 conjugation to L166P DJ-1, a mutant of DJ-1 found in PD patients
In a latter part of this paper, we described the properties of L166P DJ-1, a mutant of DJ-1 found in PD patients,17 and K130RX DJ-1, an artificial mutant possessing four mutations including K130R, in terms of sumoylation and loss of function. Both L166LP and K130RX were improperly sumoylated in vivo compared to wild-type DJ-1, to which one SUMO-1 molecule was conjugated. It is possible that this improper SUMO-1 conjugation to L166P and K130RX occurs on multiple lysine residues (multisumoylation) or on a specific lysine residue with poly-SUMO-1 (polysumoylation). It is also possible that L166P and K130RX are misfolded, resulting in sumoylation of multiple lysine residues (multisumoylation) and then abnormal aggregation. Although polysumoylation, which had been thought not to occur, has recently been reported,55, 56 it is not clear whether there was excess sumoylation of DJ-1 mutants in either case. Since K130RX lost lysine at 130, a lysine(s) other than K130 was sumoylated. Furthermore, both L166LP and K130RX were also improperly sumoylated in an in vitro reconstituted system using recombinant proteins. The crystal structure of DJ-1 has been determined by five groups, including us, and it has been shown that L166 is located in the α-helix near the C-terminal region of DJ-1 and that L166P mutation might disrupt dimer formation.57, 58, 59, 60, 61 Structure of K130RX was also thought to be drastically changed by the computer-based analysis. The protective activity of DJ-1 against hydrogen peroxide-induced cell death has been reported to be abrogated by L166P mutation.41, 49 It is therefore thought that the structures of these mutants themselves changed to allow for improper sumoylation or abnormal aggregation. Both L166P and K130RX DJ-1 were found to become insoluble. Recently, insoluble forms of DJ-1 have been reported to be colocalized with tau or α-synuclein in fibrillar inclusions in patients with Pick's disease and multiple system atrophy, suggesting that DJ-1 is related to various neurodegenerative diseases, including PD.30, 31 It would therefore be interesting to examine whether such insoluble forms of DJ-1 in patients with neurodegerative diseases are sumoylated.
Localization of K130R DJ-1 in the cytoplasm and nucleus was found to be the same as that of wild-type DJ-1, suggesting that SUMO-1 conjugation is not sufficient to determine the localization of DJ-1 in cells. Some of wild-type DJ-1 and K130R DJ-1 were also found to be localized in the mitochondria. Both L166P and K130RX DJ-1 were then found to be localized in the mitochondria. Since the mitochondrion is a major ROS-production organelle and since low activity levels of the complex 1 that reside in mitochondria have been found in PD patients (see review and references therein),62 it is interesting that improperly sumoylated DJ-1 was localized in the mitochondria. Since DJ-1 has no mitochondria-import/export sequences, wild-type DJ1 and mutants of DJ-1, especially L166P and L130RX, may associate with other protein(s) that are targeted in mitochondria. To determine the importance of different localizations of DJ-1 in cells, localization of DJ-1 in specific organelles such as mitochondria in patients with various neurodegenerative diseases, including PD, must be investigated.
K130RX is unstable and degraded, in part, by the ubiquitin–proteasome system, like L166P as described previously.24, 25, 26, 27, 28, 49 In the presence of the proteasome inhibitor MG132, a ladder above the position of the K130RX monomer was observed in polyacrylamide gel. This is likely to be a ubiquitinated form of K130RX DJ-1, and further study is needed to confirm this. Wild-type DJ-1 and its mutants were not affected by the presence of E64, an inhibitor of cysteine protease, indicating that DJ-1 is not regulated by its protease activity.
We established DJ-1-knockdown cells (D2 cells) in which siRNA targeting to the mouse DJ-1 gene was expressed under the control of the U6 promoter,49 and we found that D2 cells were highly susceptible to UV-induced apoptosis, indicating that DJ-1 plays a role in antiapoptotic activity. A cell line that harbors exogenously added L166P DJ-1 had sensitivity against UV-induced apoptosis similar to that of D2 cells. It is therefore thought that L166P DJ-1 acts in a dominant-negative fashion at least in this cell line, in which wild-type DJ-1 is present. In PD patients harboring L166P mutation of the DJ-1 gene, loss of function of DJ-1 triggers onset of disease.
It has been reported that improper sumoylation was related to the onset of polyglutamine diseases, Alzheimer's disease and Huntington's disease.63, 64, 65 Together with the results of those studies, the results of this study suggest that proper sumoylation is essential for DJ-1 to exert its full activities and that improper sumoylation makes DJ-1 insoluble, mislocalized and unstable, which trigger the onset of PD.
Materials and Methods
Cells
Human H1299, ME180, 293T, HeLa and rat 3Y1 cells were cultured in Dulbecco's modified Eagle's medium with 10% calf serum.
Construction of lysine mutants of DJ-1
Nucleotide sequences of the oligonucleotide used for PCR primers were as follows: DJ-1-K62R (+), 5′-CTGAAGATGCAAGAAAAGAG-3′; DJ-1-K62R (−), 5′-CATATGGTCCCTCTTTTCTTGC-3′; DJ-1-K62, 63R (+), 5′-CTGAAGATGCAAGAAGAGAG-3′; DJ-1-K62, 63R (−), 5′-CATATGGTCCCTCTCTTCTTGC-3′; DJ-1-K63R (+), 5′-CTGAAGATGCAAAAAGAGAG-3′; DJ-1-K63R (−), 5′-CATATGGTCCCTCTCTTTTTGC-3′; DJ-1-K130R (+), 5′-CAACACACCCTCTTGCTAGAGAC-3′; DJ-1-K130R (−), 5′-CATCATTTTGTCTCTAGCAAG-3′; T7, 5′-CCCTATAGTGAGTCGTATTA-3′; Sp6, 5′-TTTAGGTGACACTATAGAAT-3′. The first PCR was carried out on pcDNA3-FLAG-DJ-1 as a template either with DJ-1-K (+) and T7 or with DJ-1K (−) and Sp6 as primers. The products were mixed and used as templates for the second PCR with T7 and Sp6 as primers. The resultant PCR product was digested with HindIII and XhoI and inserted into the respective site of pcDNA3.
SUMO-1 conjugation to DJ-1 in vivo
Proteins were extracted from human H1299 cells with a buffer containing 0.1% NP-40, 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl and by sonication. Proteins were then immunoprecipitated with an anti-DJ-1 polyclonal antibody-conjugated agarose,1 and the precipitates were dissolved with a sample buffer containing 30 mM Tris-HCl (pH 6.8), 6% SDS, 0.006% bromophenol blue, 12% 2-mercaptoethanol and 50% glycerol, boiled for 15 min, and blotted with an anti-DJ-1 monoclonal antibody (3E8, specificity described by Yoshida et al.9) or an anti-SUMO-1 polyclonal antibody (FL-101, Santa Cruz). H1299 cells were transfected with 5 μg of FLAG-DJ-1 or its mutants, 2 μg of Ubc9-HA and 2 μg of T7-SUMO-1 by the calcium phosphate precipitation method. At 48 h after transfection, the proteins extracted from cells were immunoprecipitated with an anti-FLAG monoclonal antibody (M2, Roche), blotted with an anti-T7 monoclonal antibody (Novagen) or the anti-FLAG monoclonal antibody, and visualized by ECL (Amersham BioScience).
SUMO-1 conjugation to DJ-1 in vitro
Recombinant proteins of human SUMO-1, E1 enzyme (SAE1 and SAE2) and Ubc9 were purchased from Alexis Biochemicals. 35S-FLAG-DJ-1 and its mutant were synthesized in vitro using a TnT-Coupled reticulocyte lysate system (Promega). Reaction mixture containing 100 ng of E1 enzyme, 900 ng of Ubc9, 10 μg of SUMO-1 and 35S-FLAG-DJ-1 and its mutant was incubated at 30°C for 2 h, mixed with the sample buffer, and boiled for 5 min. The proteins were then separated on SDS-containing polyacrylamide gel and visualized by fluorography.
Focus forming and growth assays
Rat 3Y1 cells cultured in a 10-cm dish were transfected with 1 μg each of pEJ6.6, pCMV-F-DJ-1 or pCMV-F-DJ-1-mutant by the calcium phosphate precipitation method, and the medium was changed every 2 days. At 14 days after transfection, the cells were stained with Giemsa solution and the foci that had formed were counted.
Rat 3Y1 cells were transfected with 5 μg each of the above plasmids by the calcium phosphate precipitation method and cultured in the presence of 300 μg/ml of G418. At 14 days after transfection, the cells were stained with Giemsa solution and the colonies that had formed were counted.
Indirect immunofluorescence
H1299 cells were transfected with FLAG-DJ-1 or its mutant as described above. At 48 h after transfection, cells were fixed with a solution containing 4% paraformaldehyde and reacted with a combination of a mouse anti-FLAG monoclonal antibody (M2, Sigma), goat anti-GRP78 polyclonal antibody (N-20, Santa Cruz), and MitoTracker (Molecular Probe). The cells were then reacted with an FITC-conjugated anti-mouse IgG or rhodamine-conjugated anti-rabbit IgG and observed under a confocal laser fluorescent microscope.
Isoelectric focusing
H1299 cells were irradiated with UV using a UV cross linker, and proteins were extracted from the cells at various times after UV-irradiation. Proteins were then separated in the pH 5–8 range of isoelectric focusing phoresis gel or 12.5% polyacrylamide gel containing SDS, transferred onto nitrocellulose membranes, and blotted with an anti-DJ-1 polyclonal antibody or antiactin-antibody (MAB1501R, CHEMICON) as described previously.41
Establishment of a DJ-1-knockdown cell line
The nucleotide sequences of the upper strand of the oligonucleotide used for construction of an siRNA vector targeting mouse DJ-1 gene is 5′-GGATCCCGCCTTGCTAGTAGAATAAACTTCAAGAGAGTTTATTCTACTA GCAAGGTTTTTTCCAAAAGCTT-3′. After annealing oligonucleotides corresponding to the upper and lower strands of DNA, these were inserted into BamHI–HindIII sites of pRNA-U6.1/Neo (GenScript Corp.). These plasmids were transfected into mouse Flp-In™3T3 cells (Invitrogen) by the calcium phosphate precipitation method, and the cells were cultured in the medium in the presence of 200 μg/ml G418 for 14 days. The cells that were resistant to the drug were then selected, and expression of DJ-1 was examined by Western blotting with an anti-mouse DJ-1 antibody.8
Separation of soluble and insoluble forms of DJ-1
Human 293T cells were transfected with FLAG-DJ-1 or its mutant as described above. At 48 h after transfection, cells were lysed with a buffer containing 0.1% NP-40 and phosphate-buffered saline and by sonication. The supernatant faction after centrifugation was used as soluble proteins, and the precipitates were further dissolved with a sample buffer containing SDS as described above and used as insoluble proteins. These proteins were blotted with an anti-FLAG antibody.
Abbreviations
- AR:
-
androgen receptor
- PD:
-
Parkinson's disease
References
Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SMM and Ariga H (1997) DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 31: 509–513
Taira T, Takahashi K, Kitagawa R, Iguchi-Ariga SMM and Ariga H (2001) Molecular cloning of human and mouse DJ-1 genes and identification of Sp1dependent activation of the human DJ-1 promoter. Gene 263: 285–292
Hod Y, Pentyala SN, Whyard TC and El-Maghrabi MR (1999) Identification and characterization of a novel protein that regulates RNA-protein interaction. J. Cell. Biochem. 72: 435–444
Wagenfeld A, Yeung CH, Strupat K and Cooper TG (1998) Shedding of a rat epididymal sperm protein associated with infertility induced by ornidazole and alpha-chlorohydrin. Biol. Reprod. 8: 1257–1265
Welch JE, Barbee RR, Roberts NL, Suarez JD and Klinefelter GR (1998) SP22: a novel fertility protein from a highly conserved gene family. J. Androl. 19: 385–393
Wagenfeld A, Yeung CH, Shivaji S, Sundareswaran VR, Ariga H and Cooper TG (2000) Expression and cellular localization of contraception-associated protein. J. Androl. 21: 954–963
Klinefelter GR, Welch JE, Perreault SD, Moore HD, Zucker RM, Suarez JD, Roberts NL, Bobseine K and Jeffay S (2002) Localization of the sperm protein SP22 and inhibition of fertility in vivo and in vitro. J. Androl. 23: 48–63
Okada M, Matsumoto K, Niki T, Taira T, Iguchi-Ariga SMM and Ariga H (2002) DJ-1, a target protein for an endocrine disrupter, participates in the fertilization in mice. Biol. Pharm. Bull. 25: 853–856
Yoshida K, Sato Y, Yoshiike M, Nozawa S, Ariga H and Iwamoto T (2003) Immunocytochemical localization of DJ-1 in human male reproductive tissue. Mol. Reprod. Dev. 66: 391–397
Le Naour F, Misek DE, Krause MC, Deneux L, Giordano TJ, Scholl S and Hanash SM (2001) Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer. Clin. Cancer Res. 7: 3328–3335
Miura K, Bowman ED, Simon R, Peng AC, Robles AI, Jones RT, Katagiri T, He P, Mizukami H, Charboneau L, Kikuchi T, Liotta LA, Nakamura Y and Harris CC (2002) Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking- and prognosis-related molecular profiles. Cancer Res. 62: 3244–3250
Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SMM and Ariga H (2001) DJ1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 276: 37556–37563
Niki T, Takahashi-Niki K, Taira T, Iguchi-Ariga SMM and Ariga H (2003) DJBP: A novel DJ-1-binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex. Mol. Cancer Res. 1: 247–261
Taira T, Iguchi-Ariga SMM and Ariga H (2004) Co-localization with DJ-1 is essential for the androgen receptor to exert its transcription activity that has been impaired by androgen-antagonists. Biol. Pharm. Bull. 27: 574–577
Shinbo Y, Taira T, Niki T, Iguchi-Ariga SMM and Ariga H (2005) DJ-1 restores p53 transcription activity inhibited by Topors/p53BP3. Int. J. Oncol. 26: 641–648
Sekito A, Taira T, Niki T, Iguchi-Ariga SMM and Ariga H (2005) Stimulation of transforming activity of DJ-1 by Abstrakt, a DJ-1-binding protein. Int. J. Oncol. 26: 685–689
Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA and Heutink P (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299: 256–259
Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ and Wood NW (2003) The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann. Neurol. 54: 283–286
Hague S, Rogaeva E, Hernandez D, Gulick C, Singleton A, Hanson M, Johnson J, Weiser R, Gallardo M, Ravina B, Gwinn-Hardy K, Crawley A, St George-Hyslop PH, Lang AE, Heutink P, Bonifati V, Hardy J and Singleton A (2003) Early-onset Parkinson's disease caused by a compound heterozygous DJ-1 mutation. Ann. Neurol. 54: 271–274
Hedrich K, Djarmati A, Schafer N, Hering R, Wellenbrock C, Weiss PH, Hilker R, Vieregge P, Ozelius LJ, Heutink P, Bonifati V, Schwinger E, Lang AE, Noth J, Bressman SB, Pramstaller PP, Riess O and Klein C (2004) DJ-1 (PARK7) mutations are less frequent than Parkin (PARK2) mutations in early-onset Parkinson disease. Neurology 62: 389–394
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI and Nussbaum RL (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276: 2045–2047
Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y and Shimizu N (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608
Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD and Polymeropoulos MH (1998) The ubiquitin pathway in Parkinson's disease. Nature 395: 451–452
Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V, Hoogeveen A, Heutink P and Rizzu P (2003) The DJ-1L166P mutant protein associated with early onset Parkinson's disease is unstable and forms higher order protein complexes. Hum. Mol. Genet. 12: 2807–2816
Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, Carter DM, Zhu PP, Stadler J, Chandran J, Klinefelter GR, Blackstone C and Cookson MR (2003) L166P Mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin–proteasome system. J. Biol. Chem. 278: 36588–36595
Moore DJ, Zhang L, Dawson TM and Dawson VL (2003) A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J. Neurochem. 87: 1558–1567
Gorner K, Holtorf E, Odoy S, Nuscher B, Yamamoto A, Regula JT, Beyer K, Haass C and Kahle PJ (2004) Differential effects of Parkinson's disease-associated mutations on stability and folding of DJ-1. J. Biol. Chem. 279: 6943–6951
Olzmann JA, Brown K, Wilkinson KD, Rees HD, Huai Q, Ke H, Levey AI, Li L and Chin LS (2004) Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279: 8506–8515
Bandopadhyay R, Kingsbury AE, Cookson MR, Reid AR, Evans IM, Hope AD, Pittman AM, Lashley T, Canet-Aviles R, Miller DW, McLendon C, Strand C, Leonard AJ, Abou-Sleiman PM, Healy DG, Ariga H, Wood NW, de Silva R, Revesz T, Hardy JA and Lees AJ (2004) The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson's disease. Brain 127: 420–430
Rizzu P, Hinkle DA, Zhukareva V, Bonifati V, Severijnen LA, Martinez D, Ravid R, Kamphorst W, Eberwine JH, Lee VM, Trojanowski JQ and Heutink P (2004) DJ-1 colocalizes with tau inclusions: a link between parkinsonism and dementia. Ann. Neurol. 55: 113–118
Neumann M, Muller V, Gorner K, Kretzschmar HA, Haass C and Kahle PJ (2004) Pathological properties of the Parkinson's disease-associated protein DJ-1 in alpha-synucleinopathies and tauopathies: relevance for multiple system atrophy and Pick's disease. Acta Neuropathol. 107: 489–496
Heikkila R and Cohen G (1971) Inhibition of biogenic amine uptake by hydrogen peroxide: a mechanism for toxic effects of 6-hydroxydopamine. Science 172: 1257–1258
Nicklas WJ, Youngster SK, Kindt MV and Heikkila RE (1987) MPTP, MPP+ and mitochondrial function. Life Sci. 40: 721–729
Lotharius J and O'Malley KL (2000) The parkinsonism-inducing drug 1-methyl-4-phenylpyridinium triggers intracellular dopamine oxidation. A novel mechanism of toxicity. J. Biol. Chem. 275: 38581–38588
Jenner P (2003) Oxidative stress in Parkinson's disease. Ann. Neurol. 53 (Suppl. 3): S26–S36
Mitsumoto A, Nakagawa Y, Takeuchi A, Okawa K, Iwamatsu A and Takanezawa Y (2001) Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radical Res. 35: 301–310
Mitsumoto A and Nakagawa Y (2001) DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radical Res. 35: 885–893
Srisomsap C, Subhasitanont P, Otto A, Mueller EC, Punyarit P, Wittmann-Liebold B and Svasti J (2002) Detection of cathepsin B up-regulation in neoplastic thyroid tissues by proteomic analysis. Proteomics 2: 706–712
Kinumi T, Kimata J, Taira T, Ariga H and Niki E (2004) Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 317: 722–728
Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H and Mizusawa H (2003) Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem. Biophys. Res. Commun. 312: 1342–1348
Taira T, Saito Y, Niki T, Iguchi-Ariga SMM, Takahashi K and Ariga H (2004) DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5: 213–218
Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G and Wood NW (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1160
Schmidt D and Muller S (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell Mol. Life Sci. 60: 2561–2574
Seeler JS and Dejean A (2003) Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4: 690–699
Kahyo T, Nishida T and Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8: 713–718
Schmidt D and Muller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99: 2872–7287
Nishida T and Yasuda H (2002) PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 277: 41311–41317
Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F and Grosschedl R (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15: 3088–3103
Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SMM and Ariga H (2004) Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem. Biophys. Res Commun. 320: 389–397
Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA and Cookson MR (2004) The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 101: 9103–9108
Shendelman S, Jonason A, Martinat C, Leete T and Abeliovich A (2004) DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2: e362
Martinat C, Shendelman S, Jonason A, Leete T, Beal MF, Yang L, Floss T and Abeliovich A (2004) Sensitivity to oxidative stress in DJ-1-deficient dopamine neurons: an ES- derived cell model of primary parkinsonism. PLoS Biol. 2: e327
Kim RH, Peters M, Jang Y, Shi W, Pintilie M, Fletcher GC, DeLuca C, Liepa J, Zhou L, Snow B, Binari RC, Manoukian AS, Bray MR, Liu FF, Tsao MS and Mak TW (2005) DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 7: 263–273
Xu J, Zhong N, Wang H, Elias JE, Kim CY, Woldman I, Pifl C, Gygi SP, Geula C and Yankner BA (2005) The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 14: 1231–1241
Pichler A, Gast A, Seeler JS, Dejean A and Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120
Li Y, Wang H, Wang S, Quon D, Liu YW and Cordell B (2003) Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc. Natl. Acad. Sci. USA 100: 259–264
Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H and Inagaki F (2003) The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease. J. Biol. Chem. 278: 31380–31384
Tao X and Tong L (2003) Crystal structure of human DJ-1, a protein associated with early-onset Parkinson's diseasec. J. Biol. Chem. 278: 31372–31379
Wilson MA, Collins JL, Hod Y, Ringe D and Petsko GA (2003) The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. USA 100: 9256–9261
Huai Q, Sun Y, Wang H, Chin LS, Li L, Robinson H and Ke H (2003) Crystal structure of DJ-1/RS and implication on familial Parkinson's disease. FEBS Lett. 549: 171–175
Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, Park C, Kang SO, Suh PG, Lee HS and Cha SS (2003) Crystal structures of human DJ-1 and Escherichia coli Hsp31 that share an evolutionarily conserved domain. J. Biol. Chem. 278: 44552–44559
Orth M and Schapira AH (2002) Mitochondrial involvement in Parkinson's disease. Neurochem. Int. 40: 533–541
Chan HY, Warrick JM, Andriola I, Merry D and Bonini NM (2002) Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum. Mol. Genet. 11: 2895–2904
Ueda H, Goto J, Hashida H, Lin X, Oyanagi K, Kawano H, Zoghbi HY, Kanazawa I and Okazawa H (2002) Enhanced SUMOylation in polyglutamine diseases. Biochem. Biophys. Res. Commun. 293: 307–313
Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM and Marsh JL (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304: 100–104
Acknowledgements
We thank Yoko Misawa and Kiyomi Takaya for their technical assistance. This work was supported by grants-in-aid from the Ministry of Education, Science, Culture, Sports and Technology of Japan and the Ministry of Health, Labor and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Shinbo, Y., Niki, T., Taira, T. et al. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ 13, 96–108 (2006). https://doi.org/10.1038/sj.cdd.4401704
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401704
Keywords
This article is cited by
-
Mitochondrial LonP1 protease is implicated in the degradation of unstable Parkinson's disease-associated DJ-1/PARK 7 missense mutants
Scientific Reports (2021)
-
Post translational modification of Parkin
Biology Direct (2017)
-
Oxidative Modification and Its Implications for the Neurodegeneration of Parkinson’s Disease
Molecular Neurobiology (2017)
-
The protective role of DJ-1 in ultraviolet-induced damage of human skin: DJ-1 levels in the stratum corneum as an indicator of antioxidative defense
Archives of Dermatological Research (2015)
-
Sumoylation in neurodegenerative diseases
Cellular and Molecular Life Sciences (2013)