Abstract
The dynamic regulation of chromatin structure by histone post-translational modification is an essential regulatory mechanism that controls global gene transcription. The Kdm4 family of H3K9me2,3 and H3K36me2,3 dual specific histone demethylases has been implicated in development and tumorigenesis. Here we show that Drosophila Kdm4A and Kdm4B are together essential for mediating ecdysteroid hormone signaling during larval development. Loss of Kdm4 genes leads to globally elevated levels of the heterochromatin marker H3K9me2,3 and impedes transcriptional activation of ecdysone response genes, resulting in developmental arrest. We further show that Kdm4A interacts with the Ecdysone Receptor (EcR) and colocalizes with EcR at its target gene promoter. Our studies suggest that Kdm4A may function as a transcriptional co-activator by removing the repressive histone mark H3K9me2,3 from cognate promoters.
Similar content being viewed by others
Introduction
The Kdm4 (Jmjd2/Jhdm3) family is highly conserved. In many organisms, such as Drosophila, C. elegans and mice, it specifically removes di- and tri-methyl groups from histone H3 lysine 9 (H3K9) and lysine 36 (H3K36), both located at the N-terminal tail of the core histone H31,2, while in Arabidopsis, it is specific for di- and tri-methyl H3K273. Previous studies in various organisms have addressed the role of Kdm4 members in development4,5,6,7,8,9 as well as their prominent roles in human oncogenesis10,11,12,13,14,15. Kdm4-null rice displays flower morphology defects6 and Kdm4A regulates neural crest development in the chick embryo5. In C. elegans, depletion of Kdm4 as in genetically null animals or by RNAi, results in germline apoptosis and slows DNA replication, presumably due to an increase in H3K9me3 levels16. Kdm4D-null mice have increased H3K9me3 levels in the spermatids and transiently decreased testis weight where Kdm4D is specifically expressed, but exhibit no fertility defects8. The molecular mechanisms of Kdm4 in regulating gene transcription during animal development, however, remain obscure17.
In Drosophila, two closely related Kdm4 members, Kdm4A and Kdm4B (a.k.a. JMJD2(1) and JMJD2(2), respectively), have been identified and characterized18,19,20. Kdm4A loss-of-function adult flies, although viable and morphologically normal, show a male-specific shortening of life-span and aberrations in courtship behavior and the phenotype was associated with downregulation of Hsp22 and fruitless (fru)21, while overexpression of Kdm4A resulted in a global decrease of H3K36me3 and male lethality phenotype22. The second Kdm4 member, Kdm4B was shown to be upregulated by p53 upon UV irradiation and to mediate the nucleotide excision repair (NER) response by demethylating H3K9me3 in heterochromatin23. Microarray analysis of genes differentially regulated in Drosophila Kdm4A mutant larvae suggest that while its catalytic activity is important for the expression of a group of genes, a separate set of genes is regulated independently of its H3K9/K36 demethylation activity and that at Kdm4 target gene loci, Heterochromatin Protein 1a (HP1a) and Kdm4A antagonize each other in controlling gene expression22. Findings in Drosophila demonstrate an intimate regulatory interdependence between Kdm4 and HP1a. It has been shown that HP1a interacts with Kdm4A and augments its H3K36 demethylase activity in vitro19. Kdm4A appears to have distinct mechanisms for regulating euchromatic and heterochromatic target genes and it has been suggested that HP1a targets Kdm4A to its heterochromatin targets24.
In order to understand the biological roles of Kdm4 demethylases in Drosophila, we analyzed the developmental consequences of Kdm4A and Kdm4B double mutation and uncovered a phenotype consistent with a defect in the ecdysteroid hormone pathway. Further characterization on a molecular level showed that Kdm4 proteins exert transcriptional regulation of a subset of genes within this pathway and mediates H3K9 demethylation at the promoter of the Broad gene. Thus, we have identified a direct Kdm4 target gene in euchromatin as well as the essential role of Kdm4 proteins in development. Additionally, we have detected an in vivo interaction between Kdm4A and EcR, suggesting a role for Kdm4A as a transcriptional co-activator of EcR.
Results
Kdm4A and Kdm4B are biologically redundant
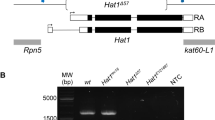
Previous studies have identified two Kdm4 family members in Drosophila based on sequence homology18,20. Although both Kdm4A and Kdm4B were found to demethylate H3K9 and H3K36 methylation in vivo, the biological functions of these demethylases remain unclear, as flies homozygous for loss-of-function mutations of either Kdm4A or Kdm4B (Figure 1A) are viable, fertile and have grossly normal morphologies (Table 1). We wondered whether the Kdm4A and Kdm4B genes were biologically redundant, i.e., that one gene product compensates for the loss of the other, thus contributing to the lack of gross abnormalities in these lines and thus tested the consequences of loss of both genes. To this end, we created a Kdm4A and Kdm4B double mutant chromosome by meiotic recombination. We found that Kdm4A,B double homozygous mutants are not viable and die in the second instar larval stage (Table S1). The lethality was rescued by expressing a Kdm4A transgene driven by Actin-Gal4. Thus, the two Kdm4 genes together are essential for viability, contrary to previous speculation that these genes were non-essential. Interestingly, one copy of either Kdm4A or Kdm4B appeared to suffice for viability and normal development (Table 1). Consistent with the idea that Kdm4A and Kdm4B are biologically redundant, in the Kdm4A homozygous loss-of-function animals, the transcript level of Kdm4B was highly upregulated and in the Kdm4B mutant genetic background, the level of Kdm4A transcripts increased (Figure 1B). Thus, in Drosophila, animal viability and development require at least one copy of either Kdm4A or Kdm4B.
Kdm4A and Kdm4B demethylate H3K9me2,3 in in vivo.
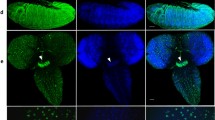
(A) A schematic representation of the Kdm4A and Kdm4B transcripts and P-element insertion site of the strains used. Coding regions are filled in with grey and non-coding regions, in white. Arrows indicate the primer pairs used for RT-PCR analysis. (B) Adult female Kdm4AKG and Kdm4BEY flies were subjected to RT-PCR analysis with primer pairs amplifying Kdm4A and Kdm4B transcripts. Note that each insertional mutation severely reduces the expression of the corresponding gene and that in each mutant there is compensatory increase in transcript levels of the other gene. (C) Total lysate was prepared from wildtype, Kdm4AKG, BEY heterozygous and homozygous mutant and single homozygous whole early second instar larvae and was subjected in SDS-PAGE, followed by blotting using antibodies against H3K9me3, H3K36me3 and H3K4me2, respectively, with Total H3 serving as a loading control. (D) Kdm4AKG, BEY homozygous double mutant or Kdm4AKG single mutant clones (lack of GFP; green) were induced in early embryos using FRT-mediated somatic recombination (see Methods), then third instar larval salivary glands were dissected and stained with anti-H3K9me2 antibody. Note the dramatically higher levels of H3K9me2 signals in mutant nuclei (GFP−) than in flanking control nuclei (GFP+). Scale bars represent 150 μ.
We then investigated the in vivo alterations in H3K9 and H3K36 levels in the single versus double Kdm4 mutants. To this end, we examined the bulk levels of H3K9 and H3K36 in second instar larvae in various mutant genetic backgrounds. Compared to the wildtype, each of the mutant animals had increased levels of trimethylated H3K9 and H3K36, consistent with the previously described roles of Kdm418,19 (Figure 1C). The double homozygous mutant animals showed a much higher augmentation of H3K9me3 and H3K36me3 compared to the double heterozygous or single homozygous lines, while having no effect on methylation of the neighboring H3K4 residue, demonstrating that both Kdm4A and Kdm4B contribute additively to the demethylation of H3K9 and H3K36 specifically in vivo. Kdm4B had a greater effect on H3K9 methylation compared to Kdm4A, consistent with previous findings18,19.
We next examined the levels of H3K9me3 by immunostaining of larval salivary glands, where the large nuclei allow for comparison of H3K9me levels on chromosomes in different genetic backgrounds. We found indeed that the H3K9me3 staining intensity was increased in Kdm4A−/− cells compared to the adjacent wildtype cells (Figure 1D). Furthermore, the Kdm4A and Kdm4B double mutant cells had an even more pronounced increase in H3K9me3 levels than did Kdm4A single mutant cells, suggesting that both Kdm4A and Kdm4B regulate H3K9 methylation in vivo.
Since we observed increased bulk levels of H3K9me3, a factor important in the recruitment of HP1a and in heterochromatin formation, we assessed heterochromatin formation in the mutant animals. At the early second instar stage, Kdm4A, B double mutants had a body and brain size comparable to those of their control siblings. However, the Kdm4A,B double mutant animals had significantly smaller and more condensed nuclei in their brains, suggesting an increased compaction of chromatin, which is consistent with the presence of a higher content of heterochromatin (Figure S1). Taken together, the above results suggest that proper H3K9 methylation and animal development require at least one copy of either Kdm4A or Kdm4B in Drosophila.
Kdm4A,B homozygous double mutation results in developmental arrest
To understand better the biological functions of Kdm4A and Kdm4B, we determined when the double homozygous mutants die and examined the phenotypic defects of the dying animals. In the first instar stage, homozygous and heterozygous double mutants were indistinguishable in their behavior and size compared to wildtype animals (Figure 2A). Differences became clear, however, at the early second instar larval stage, around the time of lethality, when the homozygous double mutants gradually lost mobility, failed to grow and deteriorated, while their wildtype or heterozygous siblings continued to grow (Figure 2B). The majority of dead homozygous double mutant animals had two sets of mouth hooks (Figure 2C) and posterior spiracles (Figure 2D), indicating retention of the first instar larval structures. In some double mutants, brown patches of deteriorated cells were seen (Figure S2). Taken together, these observations are consistent with developmental arrest and a defect in molting behavior. Disruption of the ecdysteroid pathway has been shown to cause such a phenotype25,26. Thus, our phenotypic analysis of Kdm4A and Kdm4B double mutant animals suggested that Kdm4 might regulate ecdysone pathway components.
Kdm4A,B homozygous double mutation results in developmental arrest.
Wildtype and heterozygous or homozygous Kdm4AKG, BEY loss-of-function siblings were identified by the presence or absence of GFP and were imaged at the (A) first and (B) second instar larval stages. Second instar wildtype and Kdm4AKG, BEY homozygous larvae were imaged to visualize the mouthhook (C) and posterior spiracles (D). Arrowheads point to first instar structures and arrows point to second instar structures.
Kdm4A and B regulate ecdysteroid pathway genes by promoter H3K9 demethylation
To investigate the involvement of Kdm4 in the ecdysteroid pathway, we determined whether the transcript levels of five ecdysteroid pathway genes, the well-characterized Early Puff genes, EcR, BR-C, 74EF, 75B and the Early-Late Puff gene, DHR3 were altered in the double mutants at the early second instar larval stage before they died. Indeed, four of the ecdysone-controlled genes, EcR, 75B, BR-C and DHR3 were downregulated, whereas 74EF was upregulated in Kdm4A and Kdm4B double mutant animals (Figure 3A). The ecdysone signaling pathway is organized in a hierarchical manner, with some protein products repressing their own genes or other genes and therefore it is possible that 74EF was derepressed due to the absence of other players in the cascade, as previously shown27,28,29. Thus, our results suggested that disruption of ecdysone response may be the cause of lethality.
Kdm4A, B double homozygous mutants have reduced transcript levels of ecdysteroid pathway components.
(A) Wildtype (+/+), Kdm4AKG, BEY trans-heterozygous (+/−) or double homozygous (−/−) early second instar larvae were collected at the 51 to 54 hours AEL time point. Whole larvae were utilized for RT-PCR analysis to assess changes in transcript levels of EcR, BR-C, 74EF, 75B and DHR3 ecdysteroid puff genes, with rp49 serving as an internal control. (B) The ecdysteroid pathway was activated with 1 μM 20E treatment for 1 hour in S2 cells that had been treated with dsRNA specific for Kdm4A and Kdm4B, or for GFP (control). EcR, BR-C, 74EF, 75B and DHR3 transcript levels were subjected to quanitative-PCR analysis in triplicate. Transcript levels were normalized to rp49 and then expressed as fold change over those without 20 E treatment. Error bars represent standard deviation. (C) A schematic representation of the Broad genomic locus and primers used for ChIP. The light gray boxes represent the upstream EcR Responsive region36 and a region enriched for both EcR and PolII binding (ModEncode.org). The solid line represents genomic region and the dark gray box BR-C transcript. The arrows represent primer pairs used for ChIP. (D) ChIP analysis was conducted with anti-H3K9me3 antibodies on wildtype and Kdm4AKG, BEY homozygous early second instar whole larvae staged at 51–54 AEL. At least three sets were analyzed. The Fold Enrichment method was used for calculation, with normalization to the actin5C proximal promoter primer sets. Error bars represent standard error. (E) S2 cells were treated with dsRNA specific for Kdm4A and Kdm4B, or for GFP (control), then with or without 1 μM of 20E for 1 hour and were harvested and subjected to ChIP analysis with anti-H3K9me3 antibody in triplicates. The Fold Enrichment method was used for calculation, normalized to primer pairs amplifying the actin5C proximal promoter region. Error bars represent standard error.
Despite the dramatic effects on ecdysone target genes, the Kdm4 genes did not seem to be required for transcription of all genes. The Kdm4 homozygous mutant animals did not display gross morphological defects, retaining a normal body plan and normal segment polarity. We examined the expression of Engrailed (En), a transcription factor that directs posterior compartment specification during segmentation and found that the pattern and intensity of immunostaining in second instar larval brains were similar when wild-type and mutant animals were compared (Figure S3A). We also compared the expression of En in Kdm4−/− cells compared to neighboring wild-type cells in the posterior compartment of the larval wing disc where En is specifically expressed and observed no difference (Figure S3B). Taken together, Kdm4 does not affect en expression, while being important for ecdysteroid signaling, suggesting the latter specifically requires Kdm4.
In order to investigate whether Kdm4A and Kdm4B in fact play a role in the induction of ecdysteroid target genes, EcR, BR-C, 74EF, 75B and DHR3, we knocked down both Kdm4A and Kdm4B in cultured S2 cells. By treating cells with double-stranded RNA directed against Kdm4A and Kdm4B, levels of both Kdm4A and Kdm4B transcripts were reduced by approximately 65% compared to the control (Figure S4A). Transcript levels of the ecdysone signaling components peaked one hour after addition of the hormone, 20E (Figure S4B). While knocking down both Kdm4A and Kdm4B did not significantly alter basal transcript levels of the ecdysone signaling components (Figure S4C), the increase in BR-C and DHR3 transcripts seen in control cells after hormone addition was significantly compromised (Figure 3B). Thus, Kdm4 proteins are important for the induction of at least some of ecdysone responsive genes.
Since Kdm4 demethylases also act on H3K36 and since previous studies have demonstrated a role for H3K36 methylation in alternative splicing30,31,32, we questioned whether 74EF, which did not show a difference in total transcript levels under hormone-activated Kdm4 knockdown conditions (Figure 3B), might be post-transcriptionally regulated by Kdm4. To this end, we examined the ratio of a specific exon in one splice variant of 74EF transcripts to an exon common to all splice forms and saw no significant difference between the double stranded Kdm4 control cells (Figure S4D). Although a change in splicing was not detected with this particular exon, we have not ruled out the possibility that other splicing events were affected. Nonetheless, our results are consistent with the idea that demethylation of H3K9 rather than H3K36 is important for Kdm4-mediated expression of the Ecdysone pathway genes.
H3K9 hypermethylation at gene promoters hinders transcription by recruiting HP1a33,34. Since we observed downregulation of BR-C in the early second instar larvae of Kdm4 double mutants, as well as decreased transcriptional inducibility in S2 cells when Kdm4A and Kdm4B were knocked down, we investigated whether Kdm4A and Kdm4B regulate H3K9 methylation at the promoter of the BR-C locus. Using information available from the ModEncode data set, as well as prior publications describing the EcR-responsive promoter element of BR-C35,36,37, we designed various primers across the BR-C locus (Figure 3C), as well as the actin5C locus, which severs as a control. Indeed, a significant enrichment of H3K9me3 was detected at the EcR-only and EcR and PolII enriched regions of the ecdysone responsive region of the BR-C promoter in Kdm4A and Kdm4B homozygous double mutant second instar larvae compared to wildtype (Figure 3D). Additionally, we investigated alterations at the BR-C promoter upon hormone induction in S2 cells and found that H3K9me3 levels at the EcR and PolII enriched region was significantly reduced (Figure S4E). Knocking down Kdm4A and B, however, blocked 20E-induced reduction of H3K9me3 levels at the BR-C promoter region (Figure 3E). These results suggest that Kdm4A and Kdm4B demethylate H3K9me3 specifically at the promoter of BR-C in response to ecdysone signals.
Kdm4A colocalizes and physically interacts with EcR
Having shown that Kdm4 participates in transcriptional regulation of the ecdysone pathway target genes, we next investigated how Kdm4 is involved in the functions of EcR, the major transcription factor in ecdysone signaling. To this end, we first examined whether Kdm4 is localized at ecdysone responsive genomic loci by immunostaining for transgene-expressed Kdm4A-Flag on salivary gland polytene chromosomes. We found that Kdm4-Flag staining was excluded from HP1a and DNA stain-rich regions (Figure S3), indicating that it localizes to euchromatin, consistent with previously described findings18,19. Moreover, we detected co-localiztion of Kdm4A-Flag with EcR at a number of loci, including the BR-C locus (Figure 4A, A′), consistently with our previous data demonstrating Kdm4's involvement in BR-C transcriptional regulation (Figure 3).
Kdm4A interacts with EcR in vivo.
(A) Flag-tagged Kdm4A was expressed in the salivary gland under the control of the Sgs-GAL4 driver and third instar larval polytene chromosomes were prepared. Immunostaining was performed to visualize the localization of Kdm4A-Flag (green) and EcR (red) on chromosomes, visualized by the DNA dye Toto3 (magenta). The scale bar represents 150 μm. (A′) A higher magnification of a region in A (white dashed line). The Broad (BR-C) locus is indicated with an arrowhead. Note the colocalization of EcR and Kdm4A-Flag signals at the Br-C locus. The scale bar represents 50 μm. (B) UAS-Kdm4A-Flag lines or wildtype control flies were crossed to the ubiquitous T80-GAL4 driver. Protein lysates were prepared from stationary third instar larvae and immunoprecipitation was done with an anti-Flag antibody. The immunoprecipitates and input lysate were subjected to SDS-PAGE and blotted with anti-EcR common domain, anti-Flag and anti-β-tubulin antibodies, respectively. (C) S2 cells were treated with,no 20E, or with 1 μM or 10 μM of the hormone. Transcript levels of Kdm4A and Kdm4B were measured by quantitative PCR analysis in triplicate. Error bars represent standard deviation. (D) S2 cells were co-transfected with Kdm4A-Flag and EcR-HA in the presence of 20E. The cells were fixed and immunostained with anti-Flag (green), anti-HA (red) and anti-H3K9me2 (magenta) antibodies. Note that cells expressing both Kdm4A-Flag and EcR-HA exhibit much reduced H3K9me2 signals (white arrows).
We next sought to investigate the interaction between Kdm4 and EcR in vivo using the Kdm4A-Flag transgene. Indeed, we detected their interaction in co-immunoprecipitation experiments (Figure 4B), consistent with their co-localization at the BR-C locus on polytene chromosomes (Figure 4A′). Moreover, we found that adding active ecdysone hormone (20E) to cultured S2 cells resulted in an up-regulation of both Kdm4A and Kdm4B (Figure 4C). These observations suggest that Kdm4 may be an integral, previously unidentified component of the ecdysone signaling pathway, important for the maximal biological response to hormonal peaks throughout development.
To understand the significance of the Kdm4A-EcR interaction, we tested whether binding to EcR could alter the demethylase activity of Kdm4A. To this end, we co-transfected Kdm4A-Flag and EcR-HA into Drosophila S2 cells and compared the levels of the heterochromatin mark H3K9me3 in co-transfected cells with those in singly transfected cells. We found that, while expressing EcR had no apparent effect on H3K9me3 levels, expressing Kdm4A caused a decrease in H3K9me3 levels (Fig. 4D), consistent with a previous report18. When Kdm4A and EcR were both expressed, however, the levels of H3K9me3 were further decreased in a subset of cells (Fig. 4D, arrows). These results suggest the possibility that binding to EcR might enhance the enzymatic activity of Kdm4A as an H3K9me3 demethylase.
Discussion
In this study, we have diccovered a role for Kdm4 in the transcriptional regulation of a subset of ecdysone pathway components. Furthermore, we have demonstrated an interaction between Kdm4A and EcR in vivo, providing evidence that Kdm4 demethylases may act as co-activators of EcR. Our genetic approach has allowed us to detect a previously uncharacterized, but essential, role of Kdm4 in development and to identify a direct Kdm4 target gene in euchromatin. Interestingly, Human Kdm4 members interact with the nuclear hormone receptors, Androgen Receptor (AR) and Estrogen Receptor α (ERα) and have been proposed to serve as co-activators, suggesting a molecular mechanism by which Kdm4 can act as an oncogene in prostate and breast cancers10,11,13. Kdm4B was shown to be a direct target gene of ERα, yielding a feed-forward loop for an augmented hormonal response10. Our results indicate that a similar epigenetic mechanism exists in Drosophila, where a nuclear hormone receptor requires the Kdm4 family of demethylases to remove H3K9 methylation at the promoter of a target gene10,11,13. Taken together, the Kdm4 family of demethylases may function as transcriptional co-factors required for transcriptional activation by nuclear hormone receptors.
Previous studies have shown that the Trithorax-related (Trr) H3K4 methyltransferase, the Nurf nucleosome remodeling complex component, Nurf301, the Brahma (Brm)-containing chromatin remodeler and the histone acetyltransferase CREB-binding protein (CBP) are also co-activators of EcR, indicating that activation of ecdysone pathway genes requires substantial regulation of the chromatin environment38,39,40. Since H3K4 hyper-methylation at promoters is a marker of active transcription and since H3K9 hypo-methylation also promotes upregulation of gene expression, it is feasible that synchronizing these two events would lead to more robust target gene activation. The mammalian Kdm4B (JMJD2B) forms a complex with the mixed-lineage leukemia (MLL) 2 H3K4 methyltransferase and serves as a co-activator of Estrogen Receptor α (ERα)10. The complex couples H3K9 demethylation with H3K4 methylation in order to facilitate ERα target gene activation. Similar functional cross-talk between H3K9 demethylation and H3K4 methylation has been described in S. pombe, where the Lsd1 H3K9 demethylase and the Set1 H3K4 methyltransferase were found in a complex41. Since, in Drosophila, the Nurf301 subunit, Brm and CBP were also found to interact with EcR, nucleosome remodeling may cooperate as well in the rapid and dynamic activation of ecdysone regulated genes.
Our studies of the Kdm4A and Kdm4B homozygous double mutants demonstrate a requirement for these genes in the ecdysone pathway. This observation is similar to results obtained with mutant alleles of Nurf301 and trr, two seemingly ubiquitous chromatin regulators, where specific downregulation of ecdysone signaling genes has been detected38,39. Additionally, our study is consistent with the reports that adult male Kdm4A mutants display abnormal courtship behavior and concomitant downregulated fru, a gene speculated to be a direct downstream target of EcR42,43. The specific defects in ecdysone signaling, rather than general transcription, exhibited by the double mutants indicate that either Kdm4 may not be essential for regulating all genes, or that the aberrant expression of ecdysone responsive genes is the earliest manifestation of loss of Kdm4. However, this study does not rule out the possibility that Kdm4 proteins regulate other crucial transcription factors that in turn regulate ecdysone pathway components by secondary effects. Further molecular and genomic studies are required to resolve this issue.
We have demonstrated H3K9 demethylation-dependent transcriptional activation of BR-C. It is possible however, that H3K36 demethylation also contributes to ecdysone pathway component regulation. Previous studies have shown that HP1a is recruited to developmental puffs in polytene chromosomes and that it stimulates H3K36 demethylation by Kdm4A19,44. Perhaps H3K36 demethylation in the gene body and subsequent displacement of the HDAC complex is important for transcriptional elongation or for the activation of downstream nested promoters of ecdysone pathway components. Moreover, H3K36 plays a role in exon splice choice and thus ecdysone pathway genes that produce multiple splice variants may require Kdm4 regulation30,31,32. However, our immunostaining experiments show that HP1a and Kdm4A signals are mostly non-overlapping (Fig. S4D). Thus, it seems that HP1a's involvement in the demethylase activities of Kdm4 toward H3K9 or H3K36 would have to be transient and dynamic.
In summary, we have shown that double homozygous mutants of the two Kdm4 genes in Drosophila display developmental delays and lethality, with compromised activation of ecdysone related genes. Furthermore, we have found that BR-C may be a direct target of H3K9 demethylation and that the interaction between Kdm4A and EcR may be important in transcriptional activation of BR-C. These results provide insight into the physiological functions and mechanistic roles of Kdm4 in vivo. The interaction between Kdm4 and EcR awaits further investigation. It is conceivable that EcR directs the recruitment of Kdm4A to the promoter of its target genes, or alternatively, that EcR allosterically regulates the demethylase activity of Kdm4A, allowing removal of H3K9m2,3 only upon hormone signaling (Figure 5).
Model of Kdm4 regulation of EcR target genes.
Our results lead us to propose a model in which binding of the activated Ecdysone hormone (20E) to EcR and protein-protein interaction between EcR and Kdm4, promote H3K9 demethylation at the promoters of EcR target genes. The demethylation may be a consequence of sequence specific recruitment and targeting of Kdm4 by EcR, or a stimulation by EcR of the demethylase activity of Kdm4.
Methods
Drosophila strains and DNA plasmids
P-element insertion lines of Kdm4, Kdm4AKG04636 (BL-13828) and Kdm4BEY10737 (BL-20208) were from the Bloomington Stock center (Bloomington, IN) and are described in Flybase. The UAS-Kdm4A-Flag fly strain and UAS-Kdm4A-Flag plasmids were generous gifts from Dr. Ferran Azorín (IRB Barcelona). All lines were raised at 25°C. UAS-EcR-HA plasmid was made by inserting a Not1, EcoR1 fragment containing the DNA and hormone-binding domains of EcR (cDNA from Dr. Carl Sung, University of Hawaii) into pUAST-HA.
Western blot
Total lysate was prepared for each of the indicated genotype by homogenizing an equivalent of one whole early second instar larva in 10 μl of 1X Lysis Buffer (20 mM Tris-HCL, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Sodium Pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 ug/mL leupeptin, 2 mM DTT, 25 mM Benzamidine, 1 mM PMSF, IGEPAL CA-630, 1X PIC). Samples were centrifuged at 13,000×g at 4°C, then the supernatant was boiled for 5 minutes in the presence of sodium dodecyl sulphate (SDS) sample buffer. Electrophoresis was conducted in an 8% sodium dodecyl sulphate-polyacrylamide gel (SDS-PAGE), with an equivalent of 10 μl of total lysate per lane and protein was transferred to a nitrocellulose membrane. The membrane was blocked for one hour at room temperature in TBS-T (1X Tris-buffered saline, pH 7.6 with 0.1% Triton-X) with 5% Bovine Serum Albumin (BSA) and incubated overnight at 4°C in fresh TBS-T with 5% BSA with primary antibodies as indicated. The following antibodies were utilized at 1:500 final concentration: anti-H3K9me3 (Upstate 07-442), anti-H3K36me3 (Millipore 04-801), anti-H3K4me2 (Upstate 07-030), anti-H3 C-terminus domain (Upstate 05-928). Blots were washed in TBS-T three times for 5 minutes each at room temperature. The membrane was incubated for two hours at room temperature in anti-rabbit horseradish peroxidase (HRP) conjugated secondary antibody at 1:3000 final concentration in TBS-T and washed three times for 5 min each at room temperature. An enhanced chemiluminescence kit (Pierce) was used for visualization.
Genetic mosaic analysis and immunostaining
FRT42D, Kdm4AKG04636, Kdm4BEY10737/CyO flies were crossed to hs-flp; FRT42D, ubi-GFP or hs-flp; FRT42D, arm-lacZ lines and the progeny were raised in vials with standard yeast food at 25°C, then heatshocked in a 37°C waterbath for 1 hour. Third instar larval salivary glands or wing discs were dissected in 1X PBS, pH 7.6 and fixed in 4% formaldehyde for 30 min in room temperature. The tissues were washed for 5 minutes per wash, three times, in PBS-T (1X PBS, pH 7.6 with 0.3% Triton-X), blocked in PBS-BT (1X PBS, pH 7.6 with 0.3% BSA and 0.3% Triton-X) for 30 minutes and then stained with 1:100 anti-H3K9me3 (Abcam; Cambridge, MA) or 1:100 anti-4D9 Engrailed/Invected (Developmental Hybridoma Bank, Iowa, IA)). The anti-rabbit or anti-mouse fluorophore conjugated secondary antibody was used at a 1:500 concentration and the tissues were visualized with confocal microscopy.
RT-PCR
Larvae were raised at 25°C on apple agar plates with excess yeast paste. To synchronize developmental timing, larvae were taken at the indicated hours after egg laying (AEL).
The Trizol (Invitrogen) method was used to extract total RNA from whole larvae. A Superscript III first strand synthesis kit (Invitrogen) was used for the synthesis of cDNA, by standard protocol with oligo dT to reverse transcribe poly-A tailed mRNA. Semi-quantitative PCR was conducted under the following thermocycler conditions: an initial denaturation step, 95°C (2 mins), 10°C touchdown followed by 20 cycles of 95°C (30 sec), 58°C for all primer sets (30 sec) and 72°C (1 min), followed by a final 72°C elongation period (5 min). The following sets of primer sets were utilized:
EcR Sense 5′ - AACCCGATGAGAACGAGAGCCAAA - 3′
EcR Anti-sense 5′ - TCGCGAAGAATATTGAGTCCGAGC - 3′
BR-C Sense 5′ - TCTTAAGGAAGGACTGCAGGGACT - 3′
BR-C Anti-sense 5′ - AGATGGACGACACACAGCACTTCT - 3′
74EF Sense 5′ - TGGTGCAGGGAATGATAGCCATTG - 3′
74EF Anti-sense 5′ - GGGCACCTTTCTGCATCCGAATTT - 3′
75B Sense 5′ - TCGCATCACCTGGCCATTTAGACA - 3′
75B Anti-sense 5′ - CTGTGAGTTCACCAAGGAGAAGGT - 3′
DHR3 Sense 5′ - ATTGTTGTGATGCAGCTGGTCGCT - 3′
DHR3 Anti-sense 5′ - ACCGTGTTAATCGCAACCGATGTC - 3′
74EF common exon Sense 5′ – GTGTTGCTGCTGCCTTAGGTGATT – 3′
74EF common exon Anti-sense 5′ – AATGTGGAGGAACCAGGTGCCATA – 3′
Electrophoresis was conducted using a 0.8% agarose gel.
Kdm4A and Kdm4B double stranded RNA treatment
W1118 genomic DNA or the pEGFP-C1 plasmid (Clone Tech) was used as a template for PCR reactions using the following primer sequences:
Kdm4A dsRNA Sense: TAATACGACTCACTATAGGGAAGCCGTGTTTACCCCATC
Kdm4A dsRNA Anti-sense: TAATACGACTCACTATAGGGAGATGACCTTGCGCCAATT
Kdm4B dsRNA Sense: TAATACGACTCACTATAGGGATGGGCGCCACATATGTTAT
Kdm4B dsRNA Anti-sense: TAATACGACTCACTATAGGGCTACATGGAGTCCCAGGGTG
GFP dsRNA Sense: TAATACGACTCACTATAGGGTTCTTCAAGGACGACGGCAA
GFP dsRNA Anti-sense: TAATACGACTCACTATAGGGATGTGATCGCGCTTCTCGTT
Thermocycler conditions were as follows: an initial denaturation step, 95°C (2 mins), 10°C touchdown followed by 20 cycles of 95°C (30 sec), 63°C for all primer sets (30 sec) and 72°C (1 min), followed by a final 72°C elongation period (5 min). PCR products were precipitated by adding a tenth volume of 3 M Sodium Acetate and 2.5 times volumes of 100% Ethanol and leaving at −20°C for 2 hours. The tubes were centrifuged at 13,000 rpm for 20 minutes and the pellet was washed with 70% Ethanol in DEPC-water. The tubes were again centrifuged at 13,000 rpm for 5 minutes and each pellet was re-suspended in 50 μl of DEPC-water. Transcription reactions were set up using 2 μg of purified PCR product as template per reaction (Ambion MEGA Script Kit) and incubated at 37°C overnight. After the incubation period, 5 mM final conc. of EDTA was added to each reaction and the reaction was placed at 65°C for 10 minutes and then at room temperature for 20 minutes to allow for the annealing of dsRNA. The dsRNA was then treated with DNase I and RNaseA (Promega A797D 25131005) at 1:500 dilution and incubated for 30 minutes at 37°C. The RNeasy Cleanup Kit (Qiagen) was used for dsRNA purification and the dsRNA was eluted with 50 μl of DEPC-water. S2 cells were plated in a 6-well dish, with each well containing 2 × 106 cells in complete medium. The medium in each well was then replaced with 600 μl of serum-free medium with 8 μl Cellfectin II Reagent (Invitrogen) and 20 μg of dsRNA. After 3 hours of incubation at 26°C, the medium was replaced with 1 mL of serum-free medium with 20 ug of dsRNA. Following a 2-hour incubation period at 26°C, 1 mL of complete medium was added. After 6 days, the cells from each well were divided into two separate wells. After 30 min, the medium was aspirated and replaced with 2 mL of complete medium containing 1 μM 20-hydroxy-ecdysone (Sigma).
Co-immunoprecipitation
UAS-Kdm4A-Flag or W1118 control flies were crossed to the ubiquitously expressed T80-GAL4 driver line and progeny were raised in standard yeast food at 25°C. Whole late stationary third instar larvae were utilized to prepare total lysate by homogenizing the equivalent of one larva in the presence of 20 μl of 1X Lysis Buffer (20 mM Tris-HCL, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Sodium Pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/mL leupeptin, 2 mM DTT, 25 mM Benzamidine, 1 mM PMSF, 1% IGEPAL CA-630, 1X PIC). A total of 1 μg of Anti-Flag antibody (Rockland, 401-383) was placed in 500 μL of lysate and incubated overnight at 4°C on a rocking platform. A total volume of 20 μl of Protein-A agarose beads was added and the lysate was kept on a rocking platform overnight at 4°C. The supernatant was removed and the beads were washed twice in 1X PBS for 5 minutes per wash, at 4°C. The beads were boiled for 5 min with 20 μl of SDS sample buffer and centrifuged at 13,000×g and 20 μl of the supernatant was loaded into each lane of an 8% gel for SDS-PAGE. The protein was transferred from the gel to a nitrocellulose membrane and blocked for one hour at room temperature in TBS-T (1X Tris-buffered saline, pH 7.6 with 0.1% Triton-X) with 5% BSA. The primary antibodies, anti-DDA2.7 EcR common domain (Developmental Hybridoma Bank, Iowa, IA), anti-Flag (Rockland, 401-383) and anti-β-Tubulin (Sigma 5293), were diluted to final concentrations of 1:500, 1:250 and 1:500, respectively, in TBS-T with 5% BSA and the blots were incubated at 4°C overnight. The membrane was washed in TBS-T three times for 5 minutes per wash, at room temperature, then incubated for two hours in room temperature in 1:3000 anti-rabbit horseradish peroxidase (HRP) conjugated secondary antibody in TBS-T. The membrane was washed three times for 5 min per wash, at room temperature and visualized using an enhanced chemiluminescence kit (Pierce).
Chromatin immunoprecipitation assays
Larvae were grown on apple juice agar plates at 25°C with excess yeast paste and collected at the indicated time after egg laying (AEL). Approximately 1 mg of whole larvae were crushed in PBS with a pestle. A total of 1 × 107 Schneider (S2) cells were treated with 20-hydroxy-ecdysone (Sigma) at a final concentration of 1 μM in complete medium for one hour. The larval or S2 cells were cross-linked at room temperature for 15 min with formaldehyde at a final concentration of 1%, then glycine was added to a final concentration of 0.125 mM and the samples were incubated for 5 min at room temperature. The cells were pelleted by centrifugation at 4,000 g and washed 3 times with PBS-T (1X PBS, pH 7.6 with 0.3% Triton-X). Lysis Buffer (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton-X, 0.1% Sodium Deoxycholate, 5 mM PMSF, 1X PIC) was added and the cells were sonicated 8 times with 15 second pulses and with 1 min in between pulses, with a Branson S-450 Sonifier using a setting of 40% and output of 5. The samples were centrifuged at 13,000 g for 2 min at 4°C to remove cell debris. A portion of the resulting chromatin lysate was incubated in Elution Buffer (1% SDS, 100 mM NaHCO3) at 65°C overnight and 11% of the chromatin lysate was prepared using the QIAquick PCR Purification Kit (Qiagen Cat. 28106) and was used as input. The remaining chromatin lysate was incubated overnight at 4°C in 4 μg of anti-H3K9me3 (Upstate 07-442) or an equivalent volume of water for mock treatment. Agarose-A beads were pre-blocked by overnight incubation at 4°C in 1.5 μg salmon sperm DNA per 20 μl beads, then 20 μl of the beads was added to each chromatin lysate that has been treated with antibody or control and the samples were kept on a rocking platform overnight at 4°C. The beads were washed three times for 5 min per wash at 4°C in Wash Solution (0.1% SDS, 1% Triton-X, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl pH 8.0), then 2 hours at 4°C in Final Wash Solution (0.1% SDS, 1% Triton-X, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-Cl, pH 8.0). The beads were incubated in Elution Buffer at room temperature for 20 min and at 65°C overnight to remove crosslinks. Chromatin was isolated using a QIAquick PCR Purification Kit and was then subjected to Sybr Green qPCR. The following sets of primer sets were utilized:
BR-C Upstream EcRE Sense 5′ - ATTCCGAAATGGCACTGGATCGAC - 3′
BR-C Upstream EcRE Anti-sense 5′ - TACGTAATACGCGAGGAGGTCAAC - 3′
BR-C PolII Enriched Region Sense 5′ - GAGAGCGATTCGCATTTCGCGTTT - 3′
BR-C PolII Enriched Region Anti-sense 5′ - AACCGATAAGTGTTGCGTTGTGCC - 3′
BR-C Common Exon Sense 5′ - AACTGGGTTTCGGATTGGCATTGG - 3′
BR-C Common Exon Anti-sense 5′ - TTGTCGCTGATGGAGATTCCGTTG - 3′
BR-C Common Intron Sense 5′ - TCCGCCAGGCTTGTTTGATGAACT - 3′
BR-C Common Intron Anti-sense 5′ - CACTTGCCAGGGAATCGGATTGTT - 3′
actin5C PolII Enriched Region Sense 5′ - GCACCGTGACCATCACAGCATAAA - 3′
actin5C PolII Enriched Region Anti-sense 5′ - AATTTCCTCCGCAACTGGGTGTTC - 3′
actin5C Promoter Sense 5′ - ATTCAACACACCAGCGCTCTCCTT - 3′
actin5C Promoter Anti-sense 5′ - ACCGCACGGTTTGAAAGGAATGAC - 3′
The Fold Enrichment method was used to calculate relative enrichment.
Polytene chromosome staining
UAS-Kdm4A-Flag was crossed to Sgs-GAL4 and the progeny were raised in vials with standard yeast food under uncrowded conditions at 25°C. Late third instar larval salivary glands were dissected in PBS-T (1X PBS, pH 7.6 with 0.1% Triton-X), treated for 30 seconds in Buffer 1 (3.7% formaldehyde in 1X PBS, pH 7.6 with 0.1% Triton-X), then fixed for 2 minutes in Buffer 2 (3.7% formaldehyde, 50% acetic acid) and placed on glass slides. Chromosome spreads were prepared by placing coverslips on the slides and tapping the coverslips. The slides were washed three times for 5 minutes per wash in PBS-T and blocked in Blocking Solution (1X PBS, pH 7.6 with 0.1% Triton-X, 0.1% IGEPAL CA-630, 0.1% Tween-20, 3% BSA) for 1 hour at room temperature. Anti-Flag primary antibody (Rockland 401-383; 1:500) and anti-DDA2.7 EcR common domain (Hybridoma Bank; 1:50) in Blocking Solution were added to the slides and were incubated in a humidity chamber overnight at 4°C. The slides were washed three times for 5 minutes per wash in PBS-T and treated with anti-rabbit and -mouse fluorophore conjugated secondary antibodies at a 1:500 concentration in PBS-T for 4 hours at room temperature in the humidity chamber. The slides were washed three times for 5 minutes per wash in PBS-T and the chromosomes were mounted in Mowiol with Toto-3. Confocal microscopy was used to visualize the staining.
Larval brain staining
W1118 or Kdm4AKG, Kdm4BEY double homozygous mutants were raised in vials with standard yeast food at 25°C. Second instar larval brains were dissected in 1X PBS, pH 7.6 and fixed in 4% formaldehyde for 30 min at room temperature. The tissues were washed three times for 5 minutes per wash in PBS-T (1X PBS, pH 7.6 with 0.3% Triton-X), blocked in PBS-BT (1X PBS, pH 7.6 with 0.3% BSA and 0.3% Triton-X) for 30 minutes and then stained with anti-4D9 Engrailed/Invected (Hybridoma Bank; 1:100). An anti-mouse fluorophore conjugated secondary antibody was used at a 1:500 concentration and the tissues were visualized using confocal microscopy.
Larval brain neuroblast squashes
Brains of second instar W1118 larvae or Kdm4AKG, Kdm4BEY double homozygous mutant larvae were dissected in 0.7% NaCl solution. The isolated brains were placed in a 0.7% NaCl solution with 0.25 μM colchicine for 1 hour at room temperature. The solution was then replaced with a hypotonic solution of 0.5% sodium citrate and incubated for 7 min at room temperature. The brains were placed in Solution A (1X PBS, 0.02% Triton-X, 3.7% formaldehyde) solution for 30 seconds, then in Solution B (3.7% formaldehyde, 20% acetic acid) for 3 minutes. Each brain, submerged in liquid, was placed on a slide, a coverslip was placed on top and the slide was sandwiched in between two filter papers. Using the thumb, firm pressure was applied vertically to squash the mitotic chromosomes. The slides were placed at −80°C for a few minutes and the coverslip was removed. The chromosomes were mounted in mowiol solution supplemented with 100 μg/mL of DAPI and examined using under epifluorescence microscopy.
Change history
04 March 2014
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Whetstine, J. R. et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–81 (2006).
Klose, R. J. et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–6 (2006).
Lu, F., Cui, X., Zhang, S., Jenuwein, T. & Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43, 715–9 (2011).
Loh, Y. H., Zhang, W., Chen, X., George, J. & Ng, H. H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21, 2545–57 (2007).
Strobl-Mazzulla, P. H., Sauka-Spengler, T. & Bronner-Fraser, M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell 19, 460–8 (2010).
Sun, Q. & Zhou, D. X. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A 105, 13679–84 (2008).
Black, J. C. et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Molecular cell 40, 736–48 (2010).
Iwamori, N., Zhao, M., Meistrich, M. L. & Matzuk, M. M. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod 84, 1225–34 (2011).
Ye, L. et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61 (2012).
Shi, L. et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A 108, 7541–6 (2011).
Shin, S. & Janknecht, R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun 359, 742–6 (2007).
Gray, S. G. et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem 280, 28507–18 (2005).
Wissmann, M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 9, 347–53 (2007).
Gaughan, L. et al. KDM4B is a Master Regulator of the Estrogen Receptor Signalling Cascade. Nucleic Acids Res 41, 6892–6904 (2013).
Coffey, K. et al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res 41, 4433–46 (2013).
Black, J. C. et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol Cell 40, 736–48 (2010).
Pedersen, M. T. & Helin, K. Histone demethylases in development and disease. Trends Cell Biol 20, 662–71 (2010).
Lloret-Llinares, M., Carre, C., Vaquero, A., de Olano, N. & Azorin, F. Characterization of Drosophila melanogaster JmjC + N histone demethylases. Nucleic Acids Res 36, 2852–63 (2008).
Lin, C. H. et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 32, 696–706 (2008).
Katoh, M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol 24, 1623–8 (2004).
Lorbeck, M. T. et al. The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene 450, 8–17 (2010).
Crona, F., Dahlberg, O., Lundberg, L. E., Larsson, J. & Mannervik, M. Gene regulation by the lysine demethylase KDM4A in Drosophila. Dev Biol 373, 453–63 (2013).
Palomera-Sanchez, Z., Bucio-Mendez, A., Valadez-Graham, V., Reynaud, E. & Zurita, M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem 285, 31370–9 (2010).
Lin, C. H., Paulson, A., Abmayr, S. M. & Workman, J. L. HP1a targets the Drosophila KDM4A demethylase to a subset of heterochromatic genes to regulate H3K36me3 levels. PLoS One 7, e39758 (2012).
Li, T. & Bender, M. A conditional rescue system reveals essential functions for the ecdysone receptor (EcR) gene during molting and metamorphosis in Drosophila. Development 127, 2897–905 (2000).
Schubiger, M., Wade, A. A., Carney, G. E., Truman, J. W. & Bender, M. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125, 2053–62 (1998).
Thummel, C. S. Ecdysone-regulated puff genes 2000. Insect Biochem Mol Biol 32, 113–20 (2002).
Andres, A. J. & Thummel, C. S. Hormones, puffs and flies: the molecular control of metamorphosis by ecdysone. Trends Genet 8, 132–8 (1992).
Ashburner, M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol 39, 141–57 (1974).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41, 376–81 (2009).
Fox-Walsh, K. & Fu, X. D. Chromatin: the final frontier in splicing regulation? Dev Cell 18, 336–8 (2010).
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Li, Y., Danzer, J. R., Alvarez, P., Belmont, A. S. & Wallrath, L. L. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130, 1817–24 (2003).
Stewart, M. D., Li, J. & Wong, J. Relationship between histone H3 lysine 9 methylation, transcription repression and heterochromatin protein 1 recruitment. Mol Cell Biol 25, 2525–38 (2005).
Roy, S. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–97 (2010).
Galceran, J., Llanos, J., Sampedro, J., Pongs, O. & Izquierdo, M. Transcription at the ecdysone-inducible locus 2B5 in Drosophila. Nucleic Acids Res 18, 539–45 (1990).
Chung, Y. T. & Keller, E. B. Regulatory elements mediating transcription from the Drosophila melanogaster actin 5C proximal promoter. Mol Cell Biol 10, 206–16 (1990).
Badenhorst, P. et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19, 2540–5 (2005).
Sedkov, Y. et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426, 78–83 (2003).
Kirilly, D. et al. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron 72, 86–100 (2011).
Li, F. et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 135, 272–83 (2008).
Beckstead, R. B., Lam, G. & Thummel, C. S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol 6, R99 (2005).
Dalton, J. E., Lebo, M. S., Sanders, L. E., Sun, F. & Arbeitman, M. N. Ecdysone receptor acts in fruitless- expressing neurons to mediate drosophila courtship behaviors. Curr Biol 19, 1447–52 (2009).
Piacentini, L., Fanti, L., Berloco, M., Perrini, B. & Pimpinelli, S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 161, 707–14 (2003).
Acknowledgements
We thank Dongdong Guo, Soma Nath and Kimberly Larson for technical assistance, Drs. Ferran Azorín (IRB Barcelona) and Jerry Workman (Stowers Institute) for the UAS-Kdm4A-Flag plasmids and transgenic flies, Dr. Carl Sung (University of Hawaii) for UAS-EcR plasmids and the Bloomington Drosophila Stock Center and the Vienna Drosophila RNAi Center (VDRC) for various Drosophila strains. Research in the Li lab is generously supported by grants from the National Institutes of Health and a Leukemia & Lymphoma Society Research Scholar grant.
Author information
Authors and Affiliations
Contributions
A.T. and W.X.L. wrote the main manuscript text. A.T. prepared figures 1–5, S1–2 and S4–6. P.D. and R.S. prepared Figure 4D. S.J.Y. prepared Figure S3. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Table and Figures
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Tsurumi, A., Dutta, P., Yan, SJ. et al. Drosophila Kdm4 demethylases in histone H3 lysine 9 demethylation and ecdysteroid signaling. Sci Rep 3, 2894 (2013). https://doi.org/10.1038/srep02894
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02894
This article is cited by
-
SetDB1 and Su(var)3-9 are essential for late stages of larval development of Drosophila melanogaster
Chromosome Research (2023)
-
Histone demethylase JMJD2B/KDM4B regulates transcriptional program via distinctive epigenetic targets and protein interactors for the maintenance of trophoblast stem cells
Scientific Reports (2021)
-
Mining histone methyltransferases and demethylases from whole genome sequence
Journal of Biosciences (2020)
-
Roles and regulation of histone methylation in animal development
Nature Reviews Molecular Cell Biology (2019)
-
Genome-wide Kdm4 histone demethylase transcriptional regulation in Drosophila
Molecular Genetics and Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.