Abstract
The Lepidopteran pest of tomato, Tuta absoluta, is native to South America and is invasive in the Mediterranean basin. The species' routes of invasion were investigated. The genetic variability of samples collected in South America, Europe, Africa and Middle East was analyzed using microsatellite markers to infer precisely the source of the invasive populations and to test the hypothesis of a single versus multiple introductions into the old world continents. This analysis provides strong evidence that the origin of the invading populations was unique and was close to or in Chile and probably in Central Chile near the town of Talca in the district of Maule.
Similar content being viewed by others
Introduction
The tomato borer, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae), is a serious pest species native to South America that recently became a major threat to tomato production in the Mediterranean Basin and may become a problem in most countries of Africa and Eurasia1,2. Tuta absoluta was first observed outside its native range in Eastern Spain in 2006 and can now be found throughout Southern Europe, North Africa and the Middle East1,2. Precise knowledge of the source population of T. absoluta invading the Mediterranean basin is required to help develop strategies to control this species3. Such knowledge is currently lacking. The tomato borer was first observed outside South America in Eastern Spain in 2006 and two years later in North Africa. This suggests a single introduction point in Southern Europe followed by a geographical expansion in Southern Europe and North Africa. However these historical records do not reveal the identity of the source population in South America. Indirect methods based on population genetics are needed to infer the invasion routes of T. absoluta. Here, we analyzed the genetic variability of samples collected in South America, Europe, Africa and the Middle East to infer precisely the source of the T. absoluta populations invading the Mediterranean Basin and to test the hypothesis of a single versus multiple introductions into the old world continents.
Results
Many samples were not at Hardy-Weinberg equilibrium (see Supplementary Table S1). These results together with the difficulty to unambiguously score microsatellite genotypes – a well-known phenomenon in Lepidoptera4 – suggested the presence of frequent null alleles. Analyses were performed with and without the 5 loci (T437, T350, T378, T482, T458) that accounted for most disequilibria. The results presented here were obtained with all 12 loci that were analyzed and those obtained with the 5 most problematic loci were qualitatively the same. Pairwise FST values and genotypic differentiation tests (see Supplementary Table S2) showed that samples from the native area were strongly and significantly differentiated between different countries (with FST values ranging from 0.14 (Argentina vs Chile) to 0.31 (Argentina vs Venezuela) and p < 10−5). Samples from the same country in South America were not significantly differentiated with the notable exception of Southern and Northern Chile that were differentiated from Central Chile (with FST of 0.075 and 0.042, respectively). Most samples from the invaded areas in Africa, Europe and Asia were moderately differentiated with mean FST values of 0.024, except the samples from South-Eastern Spain which displayed a high level of differentiation from all the other samples from the Mediterranean basin (with mean FST values of about 0.09, Supplementary Table S2). The intra population genetic variation measured with the microsatellites was moderate both in the native and invaded areas (Supplementary Table S1). Interestingly the mean number of alleles and Nei's diversity were larger in the samples from Africa, Europe and Asia that in the samples from most of South America, with the exception of the samples from Chile.
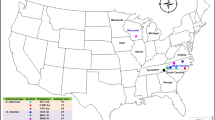
Both Bayesian clustering and a Neighbor-joining (NJ) tree grouped the samples of invasive T. absoluta into a single cluster (Figure 1 and Figure 2). This strongly suggests a single introduction of T. absoluta in the Mediterranean Basin. The Bayesian clustering analysis grouped the invasive population samples together with the samples from Chile for K = 2 and K = 3 and isolated the samples from outside South America for K = 4 (Figure 1 and Supplementary Figure S1). In addition, all the samples of the invaded area displayed minimum values of pairwise FST with the samples from central Chile to which the maximum mean assignment likelihood, Li→s, of each African, Asian and European sample corresponded (Figure 3). Moreover the NJ tree grouped the invasive population samples together with the samples from Central Chile (Figure 2). These results all suggest that Chile and particularly Central Chile, is the most probable source location of T. absoluta invading the Mediterranean Basin.
Geographic locations and Bayesian genetic clustering of the genotyped population samples of Tuta absoluta.
Note: Bar plots of the individual genetic clustering obtained from STRUCTURE (Pritchard et al. 2000) are given for K = 2 to 5 clusters. Each vertical line represents an individual and each colour represents a genetic cluster. Individuals are grouped by population sample (names at the bottom of the figure), country, continent and type of area (names at the top of the bar plots). The map was modified from the base map freely available for non-commercial use at http://www.histgeo.ac-aix-marseille.fr. In the map, the length of the horizontal black bars is 2000 km as measured at the Earth's Equator.
Mean pairwise FST (Weir & Cockerham, 1984) between invading population samples and each sample of the native area (line) and mean assignment likelihood of invading population samples to each sample of the native area (bar plots).
The vertical thin bars are the standard deviations of the parameter considered.
The results of the Approximate Bayesian Computation (ABC) analyses are shown in Table 1. These results clearly indicate that Chile is the true source of the population of T. absoluta invading the Mediterranean Basin with posterior probabilities >0.9 and non overlapping confidence intervals. Low type I and mean type II errors were obtained for both prior sets (Table 1). When Chile was removed from the analyses, the scenario with a non-sampled “ghost” South American population was selected with posterior probability >0.97 (details not shown) confirming Chile as the actual source of the invading populations of T. absoluta. Complementary analyses considering the North, Central and Southern part of Chile as 3 putative sources in addition to the clusters of Colombia and Argentina and the ghost population were performed (Supplementary Figure S2). They clearly revealed that Central part of Chile is the most probable source of invasive T. absoluta populations (Table 1), with posterior probabilities of 0.87 and 0.67 for the first and second sets of priors and samples respectively. Again, low type I and mean type II errors were obtained for both prior sets (Table 1).
Discussion
The main result of this study is that the origin of the invading populations around the Mediterranean was identified as unique and was in or close to Chile and probably Central Chile in the district of Maule. Within this region, T. absoluta displayed very weak genetic structure so that it was not possible to infer the origin of invasive populations at a finer geographical scale. The absence of genetic structure in this region, as determined by the use of neutral genetic markers, indicates that the region surroundings Talca contains a single population of T. absoluta. The choice of new natural enemies (e.g. parasitoids) for possible use in biological control should take into account this new finding concerning the origin of the invasive population T. absoluta in the Mediterranean, as suggested by Roderick & Navajas5.
The second important conclusion of this study is that the native population of T. absoluta in South America is far from genetically homogeneous. Substantial genetic differentiation was found between northern and southern regions of South America, with more than 20% of the allele frequency variance found between southern Chile and the group of Venezuela and Colombia. Such a high level of differentiation is also found over smaller distances, for instance between Argentina and Chile or between Venezuela and Colombia. Such strong genetic structure in the native area does not usually facilitate the precise inference of the source of invasive populations because it requires an extensive sampling scheme.
The third main result was that there was an almost complete absence of genetic structuring in the invaded areas, from southern Spain to Israel and from Israel to Morocco. This genetic homogeneity over space was measured using hyper variable microsatellite markers and therefore was probably not the consequence of a low power of analysis. Instead, it very probably corresponds to a single introduction in Africa or Spain followed by an expansion without noticeable a demographic bottleneck, which would have led to genetic differentiation through space. The same situation has already been found in other recent insect invasions, such as the Asian ladybeetle Harmonia axyridis expansion in France6, the western corn rootworm Diabrotica virgifera virgifera expansion in North America and Central Europe7 and the Colorado potato beetle8.
Methods
We collected samples from various regions in South America and from the invaded area in North Africa, Europe and Asia (Figure 1, Supplementary Table S1). T. absoluta larvae were collected on tomato plants in greenhouses or in open field and stored in ethanol (>90%) prior to DNA extraction. Total genomic DNA of each sampled individual was extracted using the DNeasy Tissue Kit (Qiagen) following manufacturer's instructions. The genotypes of 966 individuals were obtained at 12 microsatellite markers (T454, T425, T437, T350, T235, T310, T271, T426, T478, T378, T482, T458)9. Intra and inter sample variability statistics, including the mean number of alleles per locus, Nei's diversity10 and pairwise FST values11, were computed using Genepop12 (ver. 4). Hardy-Weinberg (HW) and genotypic differentiation tests were performed using Fisher exact tests implemented in Genepop.
The most probable source population of the European and African invasive populations was investigated as follows7. 1) First, we analyzed the pair-wise FST values between each invasive population sample and each South American sample. 2) We then computed the mean individual assignment likelihood13 (denoted Li→s) of each invading population sample i, to each possible South American source population using GENECLASS214 (ver. 2.0). The most probable source of a target invasive population's sample i was determined as the South American population whose sample displays the minimum corrected FST values with i and the maximum mean individual assignment likelihood of i. 3) We also plotted a neighbor joining (NJ) tree15 based on the genetic distance described by Cavalli-Sforza & Edwards16 using the POPULATIONS software version 1.2.30 (http://bioinformatics.org/~tryphon/populations/). It is expected that the source of a target invasive population's sample i is located in close proximity to i in the tree. 4) Finally, a Bayesian clustering analysis was performed using the STUCTURE software17 with K, the number of clusters considered varying from 1 to 10. For each value of K, an admixture model with correlated allele frequencies, the LocPrior option, 20 runs per K, 106 iterations for the MCMC and 2 × 105 iterations for the burn-in period were used. The most probable source of each target invasive population's sample i was determined as the population whose samples were the last ones to still cluster with i with increasing values of K. The most likely value of K was determined using the method of Evanno et al.18 and by eye, by examining the geographical coherence of the clustering for increasing values of K. The four above-mentioned analyses were performed on samples treated individually, i.e. without pooling samples, because frequent significant genotypic differentiation tests were found (see Supplementary Table S2).
An Approximate Bayesian Computation Analysis19 (ABC) was carried out with DIY ABC20 to measure our confidence in the source population inference. ABC is a model-based Bayesian method allowing posterior probabilities of historical scenarios to be computed, based on genetic (here, the genotypes of the samples at the 12 microsatellites) and historical data (1st observation dates of the samples) and on historical and genetic parameters priors (Supplementary Table S3, Supplementary Figure S2). We contrasted 4 historical scenarios differing by the actual source population of the Mediterranean invading populations (Supplementary Figure S2). The putative source populations were the 3 South American clusters (Venezuelan/Colombian, Argentinean and Chilean clusters) obtained from the STRUCTURE analysis described above (Figure 1) plus a non-sampled “ghost” South American population, modeling the case where the actual source population in South America had not been sampled. In each scenario the four South American populations diverged independently from an ancestral population with transitory reduction in population size at time ti (i = 1, 2, 3, 4). The analyses were conducted twice, with different parameter prior distributions and with 2 different sets of samples representative of the STRUCTURE clusters. Spa_cas and Mar_lar (see Supplementary Table S1) samples were chosen because they were large enough (>25 individuals) and they were sampled closest to the first observation point in Europe and Africa respectively1. The other samples representative of the STRUCTURE clusters were chosen randomly among the samples, with more than 25 individuals, belonging to each cluster.
References
Desneux, N. et al. Biological invasion of European tomato crops by Tuta absoluta: ecology, geographic expansion and prospects for biological control. J. Pest. Sci. 83, 197–215 (2010).
Desneux, N., Luna, M. G., Guillemaud, T. & Urbaneja, A. The invasive South American tomato pinworm, Tuta absoluta, continues to spread in Afro-Eurasia and beyond – the new threat to tomato world production. J. Pest. Sci. 84, 403–408 (2011).
Estoup, A. & Guillemaud, T. Reconstructing routes of invasion using genetic data: why, how and so what? Mol. Ecol. 19, 4113–4130 (2010).
Sinama, M. et al. Challenges of microsatellite development in Lepidoptera: Euphydryas aurinia (Nymphalidae) as a case study. Eur. J. Entomol. 108, 261–266 (2011).
Roderick, G. K. & Navajas, M. Genes in new environments: genetics and evolution in biological control. Nat. Rev. Genet. 4, 889–899 (2003).
Lombaert, E. et al. Rapid increase in dispersal during range expansion in the invasive ladybird Harmonia axyridis. J. Evol. Biol. 27, 508–517 (2014).
Ciosi, M., Miller, N. J., Toepfer, S., Estoup, A. & Guillemaud, T. Stratified dispersal and increasing genetic variation during the invasion of Central Europe by the western corn rootworm, Diabrotica virgifera virgifera. Evol. Appl. 4, 54–70 (2011).
Grapputo, A., Boman, S., Lindstroem, L., Lyytinen, A. & Mappes, J. The voyage of an invasive species across continents: genetic diversity of North American and European Colorado potato beetle populations. Mol. Ecol. 14, 4207–4219 (2005).
A'Hara, S. W. et al. Permanent Genetic Resources added to Molecular Ecology Resources Database 1 August 2011–30 September 2011. Mol. Ecol. Res. 12, 185–189 (2012).
Nei, M. Molecular Evolutionary Genetics (Columbia University Press, New York, 1987).
Weir, B. S. & Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Rousset, F. GENEPOP ' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Res. 8, 103–106 (2008).
Rannala, B. & Mountain, J. L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. 94, 9197–9201 (1997).
Piry, S. et al. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539 (2004).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Cavalli-Sforza, L. L. & Edwards, A. W. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Gen. 19, 233 (1967).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Beaumont, M. A., Zhang, W. & Balding, D. J. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002).
Cornuet, J. M. et al. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719 (2008).
Acknowledgements
We thank the “Santé des Plantes et Environnement” division of INRA for providing financial support and Alexandra Auguste for administrative and technical support. We also thank Barbara Devotto, Wilson Barros and Lucia Alarcon who helped with sample acquisition in Chile.
Author information
Authors and Affiliations
Contributions
T.G., A.T. and N.D. conceived the sampling scheme and the experiments, T.G., E.L. and A.B. analyzed the data, I.L. and A.B. performed the experiments. N.D., M.R., E.T., A.T. and L.N. organized the sampling. All authors participated to the writing of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Guillemaud, T., Blin, A., Le Goff, I. et al. The tomato borer, Tuta absoluta, invading the Mediterranean Basin, originates from a single introduction from Central Chile. Sci Rep 5, 8371 (2015). https://doi.org/10.1038/srep08371
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08371
This article is cited by
-
Reconstructing the biological invasion of Tuta absoluta: evidence of niche shift and its consequences for invasion risk assessment
Journal of Pest Science (2024)
-
Combined use of Trichoderma harzianum and Clonostachys rosea to manage Botrytis cinerea infection in tomato plants
European Journal of Plant Pathology (2023)
-
Nonreproductive effects are more important than reproductive effects in a host feeding parasitoid
Scientific Reports (2022)
-
Spatio-temporal distribution of Tuta absoluta (Meyrick 1917) (Lepidoptera: Gelechiidae) from Pakistan
International Journal of Tropical Insect Science (2022)
-
Resistance, hybrid vigor, genetic diversity, and toxicity of chemical constituents of tomatoes to Tuta absoluta (Lepidoptera: Gelechiidae)
Arthropod-Plant Interactions (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.